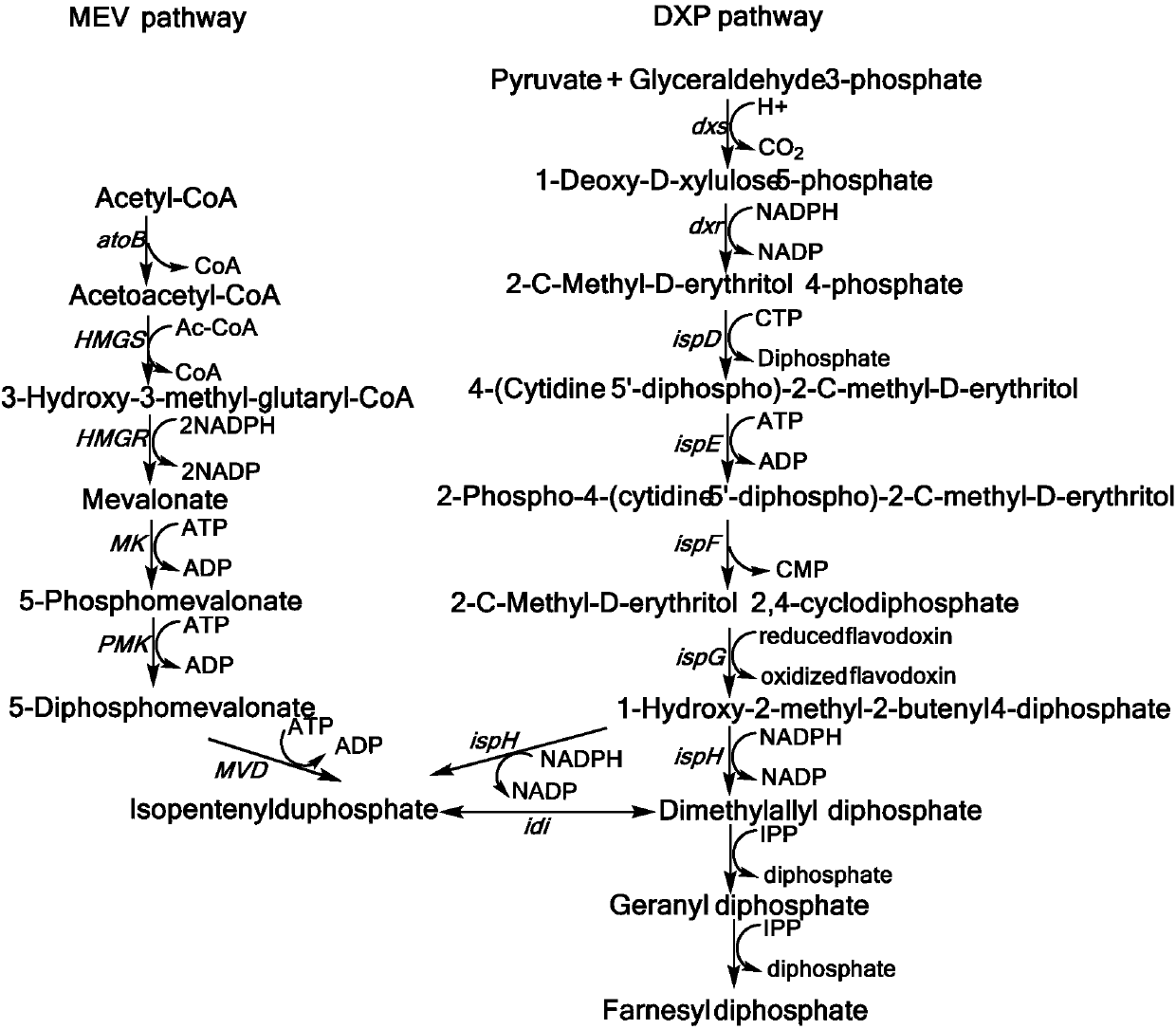
Mevalonic acid pathway tuned by TIGR
A pathway and sequence technology, applied in the field of zeaxanthin production, can solve the problems of uncoordinated expression, uncoordinated expression between genes, accumulation of intermediate products, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0116] Construction of the MEV upstream pathway regulated by embodiment 1 TIGR
[0117] 1) Using F1 / F2 as primers, Escherichia coli DH5 The genome is used as a template, the atoB gene is amplified by PCR, and connected to the pBAD33 vector to obtain pBAD33-atoB;
[0118] 2) Using F3 / F4 as primers and the Saccharomyces cerevisiae genome as a template, PCR amplifies the HMGS gene and connects it to the pMD18-T vector to obtain pMD-HMGS;
[0119] 3) Using F5 / F6 as primers and Saccharomyces cerevisiae genome as a template, PCR amplifies the tHMGR gene, and connects it to the pMD18-T vector to obtain pMD-tHMGR;
[0120] 4) PCR amplification with F7 / F8 as primers and pMD-HMGS as template; PCR amplification with F9 / F6 as primers and pMD-tHMGR as template;
[0121] 5) Using F7 / F6 as primers and the two PCR fragments recovered from the gel in the previous step as templates, amplify by PCR, connect to the SmaI / PstI space of pBAD33-atoB, and obtain pBAD33-MevTTIGR.
[0122] 6) Digest...
Embodiment 2
[0141] Example 2 MEV downstream pathway pBAD24-MevBIS
[0142] 1) Using F10 / F11, F12 / F13, and F14 / F15 as primers and Saccharomyces cerevisiae genome as a template, PCR amplifies ERG12, ERG19, and ERG8 genes, and connects them to pBAD24 to obtain pBAD24-MevB.
[0143] SEQ ID NO.14
[0144] F10:CTAGCTAGCTTTCGGAATTAAAGGAGCATCAAATATGTCTCTGCCGTTCCTG(NheI)
[0145] SEQ ID NO.15
[0146] F11:CTACCCGGGAAACTCGAGTTATGAAGTCCATGGTAAATTCG(SmaI / XhoI)
[0147] SEQ ID NO.16
[0148] F12:GATCCCGGGTTTCGGAATTAAAGGAGCATCAAATATGACCGTTTACACGGCATCC(SmaI)
[0149] SEQ ID NO.17
[0150] F13: TGCCTGCAGCCAATCGATTTATTTCTTTGGTAGACCAG (PstI / ClaI)
[0151] SEQ ID NO.18
[0152] F14: ATTCTCGAGAAAAGGGCCCTTTCGGAATTAAAGGAGCATCAAATATGTCTGAGCTGCGTGCCTTCTCTGCCCCAGGT(XhoI / ApaI)
[0153] SEQ ID NO.19
[0154] F15: ATTCCCGGGAAAAACTAGTTTTATTTATCAAGATAAGTTTCCGGA(SmaI / SpeI)
[0155] 2) Using F16 / F17 as primers and the Escherichia coli genome as a template, PCR amplifies the idi gene, connects it to pMD-18T, an...
Embodiment 3
[0165] Embodiment 3 TIGR regulates the construction of MEV downstream pathway
[0166] 1) Insert the following TIGR sequence into pBAD24-MevBIS to obtain pBAD24-MevBTIGRIS
[0167] The TIGR sequence between ERG12 and ERG8 is SEQ ID NO.3:
[0168]TIGR3:5'-GCCTAGCAAGATCTCCTGATCCCGGTGCGCGACCACCCGGACATCTGCATAGTCTGGGTGGATCAGGTACACTTACACTTGCCTTGAATTTACAGTATTTCAGTTACCGCTCTATCCTTATCCTTATCCGCTCAAGATAACCGGATACCGGCCCGATCGGTACCGCAGATACTGAATCC-3'
[0169] The TIGR sequence between ERG8 and ERG19 is SEQ ID NO.4:
[0170] TIGR4:5'-GCCTAGCAAGATCTCCTGATCCACCTTTGATGGCTAGAAAAATTAAGCTGCGGACATCTGCATAGTCTGGGCCAGTCTGAGGACTGGCGGATCAGGGCCTTGAATTTACAGTATTTAATGAACTAGCGTTCCGAGTGCATGCCTTATCCGCTCAAGACATGCACTCGGAACGCATCTAGGGTACCGCAGATACTGTATCC-3'
[0171] The above TIGR sequence can be fully synthesized or obtained by screening the library method reported by Pfleger et al. (Nature Biotechnology 2006, 24:1027-1032).
[0172] 2) pBAD24-MevBTIGRIS was digested with NheI / PstI, and connected to the correspond...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com