Application of long-chain non-coding RNA RP11-224O19.2 inhibitor
A long-chain non-coding, inhibitor technology, applied in the field of molecular biology, can solve the problem that the function of RP11-224O19.2 has not been reported yet.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Design and screening of siRNA targeting RP11-224O19.2
[0034] 1. Design and synthesis of siRNA targeting RP11-224O19.2
[0035] The siRNA targeting RP11-224O19.2 and its corresponding control siRNA (negative control siRNA) were designed by us. The sequence is shown in Table 2, and then we commissioned a company to synthesize the double-stranded siRNA sequence.
[0036] 2. Experimental grouping and cell transfection
[0037] Experimental grouping: cells were divided into 3 groups according to different treatment conditions. The experimental group (RP11-224O19.2-siRNA group) was HepG2 cells transfected with siRNA with high interference efficiency; the negative control group (Negative control siRNA group) was HepG2 cells transfected with negative control siRNA; blank group (Control group) is HepG2 cells without any treatment.
[0038] The routine culture operation of HepG2 cells is as follows:
[0039] 1) Cell recovery: Liver cancer HepG2 cells were preserved...
Embodiment 2
[0074] Example 2 siRNA targeting RP11-224O19.2 for the treatment of liver cancer
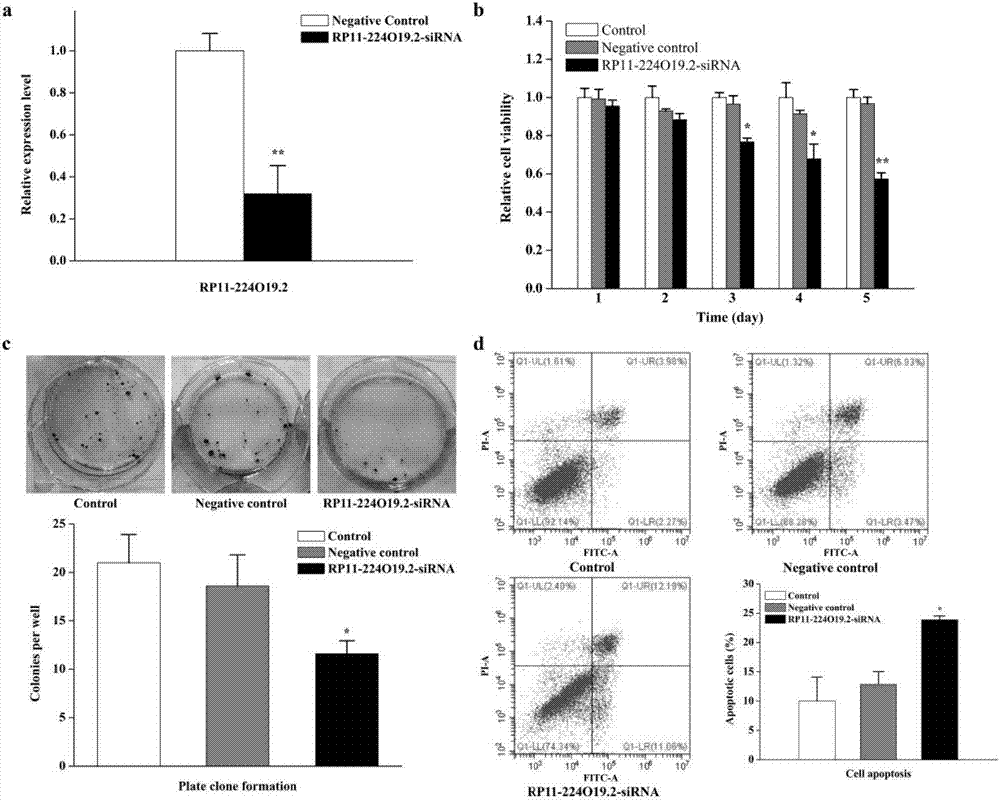
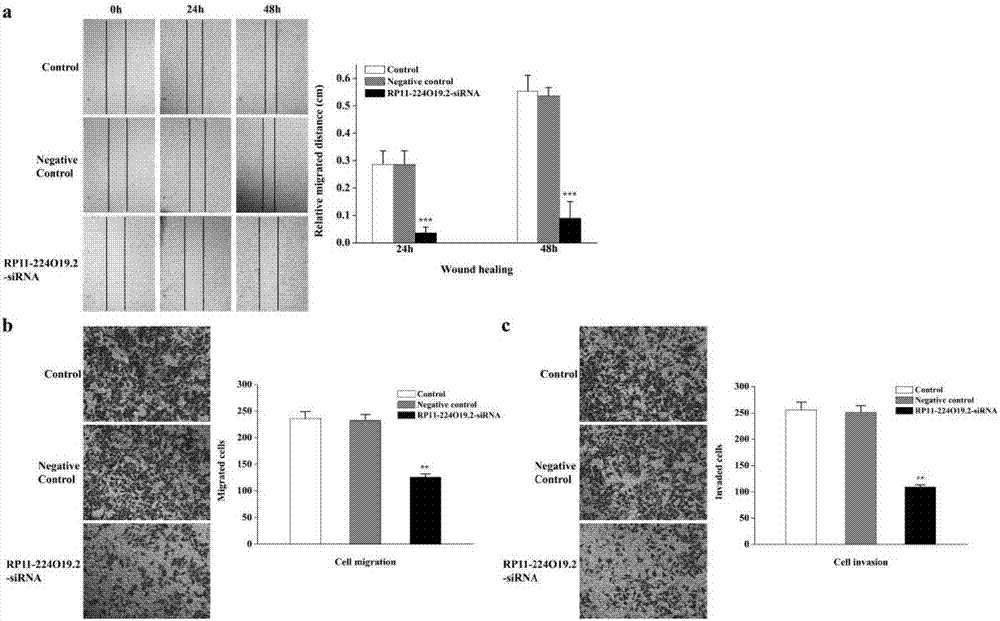
[0075] In this experiment, the expression of RP11-224O9.2 was inhibited by transfecting siRNA to verify its effect on liver cancer.
[0076] 1. Effects of inhibiting RP11-224O19.2 on the proliferation, colony formation and apoptosis of liver cancer cells
[0077] 1. CCK8 assay to detect the proliferation of HepG2 cells
[0078] The experimental steps are as follows:
[0079] 1) Plate the HepG2 cells of the transfected experimental group and the negative control group and the HepG2 cells of the untreated blank group in a 96-well plate at a plating density of 3000 cells / well, and set up a 6-well experimental group for each 96-well plate , 6-well negative control group and 6-well blank group, another 6 wells were set up for zero adjustment only with medium, and a total of 5 96-well plates were inoculated. After the plating was completed, the 96-well plates were placed in the incubator for cultiva...
Embodiment 3
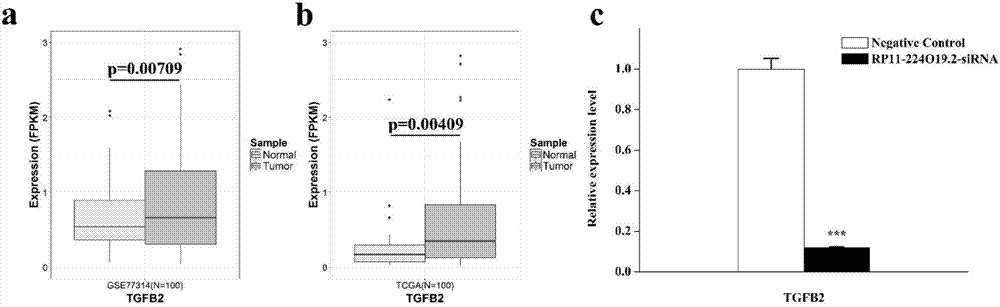
[0151] Example 3 The relationship between the expression level of RP11-224O19.2 and liver cancer
[0152] Analysis of the expression of RP11-224O19.2 in liver cancer:
[0153] The data analysis method is as follows: All experimental data were analyzed using SPSS 19.0 software (IBM SPSS), and P<0.05 between groups calculated by student's t-test was used as the statistical difference threshold, and the drawing was completed with Origin 8.0 and GraphPadPrism 5 software. According to the fold change of the expression level of RP11-224O19.2, 50 clinical cases of liver cancer were classified. The correlation analysis between the fold change of the expression level of RP11-224O19.2 and the clinicopathological parameters was completed by SPSS19.0 software. The Mann-Whitney U test method in the non-parametric test was used for the analysis among three samples, and the Kruskal-Wallis test method in the non-parametric test was used for the analysis among the three samples, and P<0.05 was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com