RNA guided eradication of human jc virus and other polyomaviruses
A polyoma virus, virus technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, and resistance to vector-borne diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0115] Example 1: Materials and methods
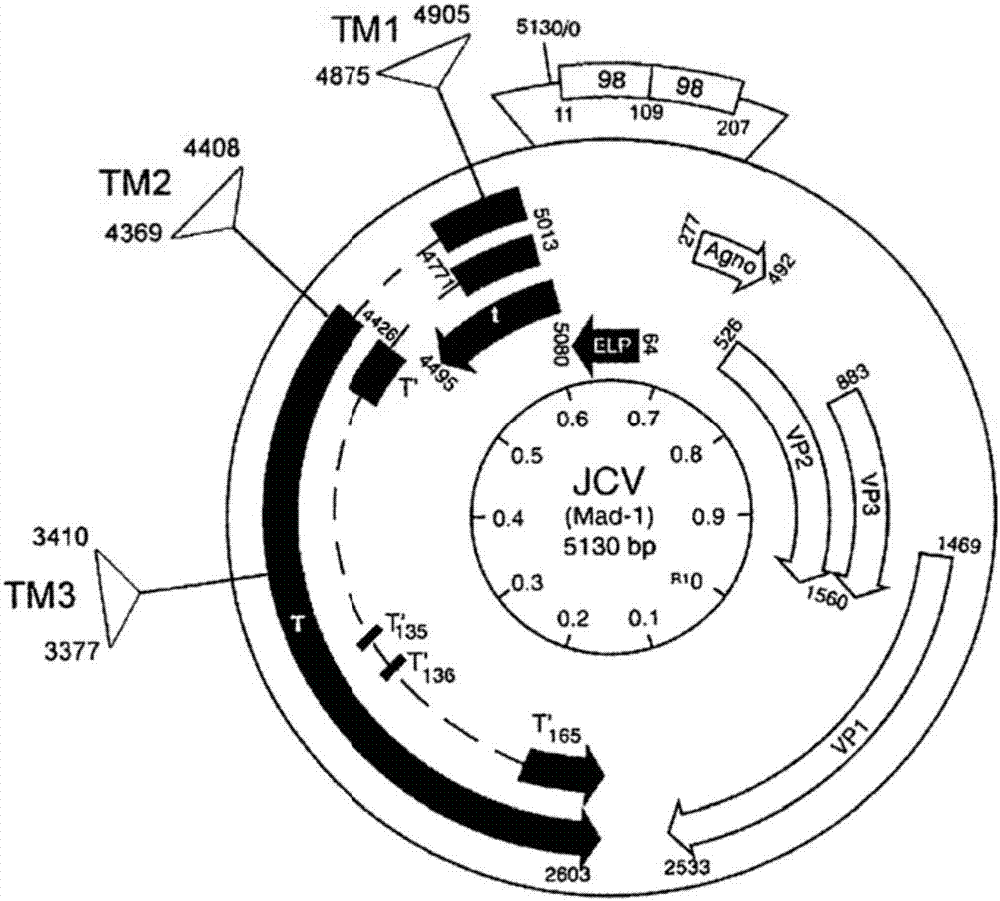
[0116] Using CRISPR / Cas9 technology to detect the effect of Cas9 and gRNA on targeting T-antigen and its function. Design of guide RNA for CRISPR / Cas9 targeting JCV as in Figure 1A and 1B shown.
[0117] cell culture . The human oligodendroglioma cell line TC620 (Wollebo et al., 2011) and SVG-A, which is derived from primary human fetal glial cells transformed with the original defective SV40 expressing SV40 T-Ag Cell line (Major et al., 1985), TC620 and SVG-A were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), as previously described (Wollebo et al., 2011). HJC-2 is a JCV-induced hamster glioblastoma cell line expressing JCV T-Ag (Raj et al., 1995). BsB8 is a mouse cell line derived from tumors of cerebellar neuroectodermal origin generated in transgenic mice expressing the JCV early protein T-Ag (Krynska et al., 2000). SVG-A cells expressing Cas9 and JCV T-antigen gRNA ...
Embodiment 2
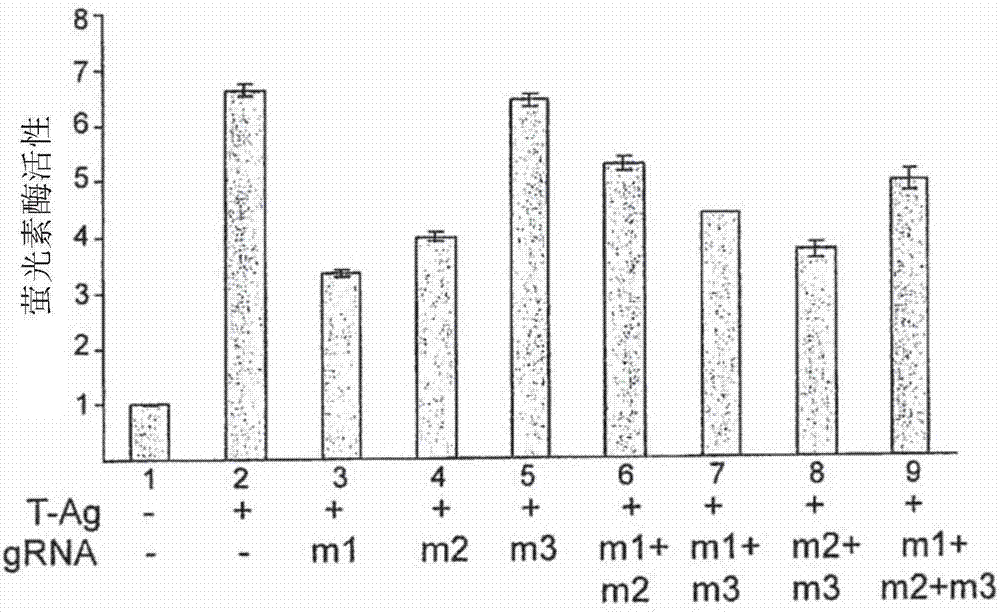
[0133] Example 2: Expression of gRNA targeting T-Ag reduces the expression of T-Ag and the expression of T-Ag-stimulated JCV late genes in TC620 cells transfected with T-Ag and Cas9
[0134] In the first set of experiments, it was determined whether the combination of Cas9 with various gRNAs targeting TM1, TM2, and TM3 could suppress T-antigen expression in the human oligodendrocyte cell line TC620. In these experiments, gRNA m1 was complementary to the TM1 target sequence of SEQ ID NO:1; gRNA m2 was complementary to the TM2 target sequence of SEQ ID NO:5; and gRNA m3 was complementary to the TM3 target sequence of SEQ ID NO:9.
[0135] Western blot analysis results from transfection of TC620 cells with plasmids expressing T-antigen alone or in combination with Cas9, and / or in combination with T-Ag expression plasmids targeting gRNA are shown in Figure 2A middle. As shown, the presence of Cas9 together with ml or m2 gRNA significantly reduced the level of T-Ag production in ...
Embodiment 3
[0137] Example 3: Clonal derivatives of SVGA expressing Cas9 and gRNA targeting T-Ag have reduced ability to support JCV infection
[0138]To investigate the effect of gRNA-guided Cas9 on JCV infection, experiments were performed with the SVG-A cell line. This line supports viral gene expression and also allows full productive viral lytic infection. First, a stable clonal cell line was established from SVG-A cells expressing Cas9 or Cas9+gRNA ml. In these experiments, gRNA ml was complementary to the TM 1 target sequence SEQ ID NO:1. Three independent clones were selected and used for JCV infection for 7 days at MOI = 1.0. Viral infection was assessed by Western blot analysis for the presence of viral capsid protein, VP1 and accessory protein, Agno protein, with α-tubulin as a loading control. Quantitative PCR (Q-PCR) was performed simultaneously to determine the level of viral DNA in the culture medium as a marker of DNA replication and thus of virus production. Such as ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com