Molecular marker closely linked with millet tillering character, primer and application
A technology of molecular markers and millet, applied in the field of molecular biology, can solve the problems of undiscovered molecular markers, etc., and achieve the effect of accelerating the process of genetic selection and improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
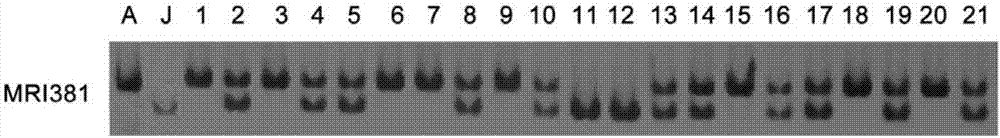
[0034] Example 1: Obtaining method of molecular marker MRI381
[0035] The female parent of the tillering trait Ai Ninghuang was used as the female parent; the non-tillering trait Jingu 21 was used as the male parent, and the F 1 , single plant F 1 543 F2 segregating populations were obtained by selfing. Using hole sowing method, the F 2 Planted in the experimental field of Millet Research Institute of Shanxi Academy of Agricultural Sciences, the row length is 6m, the row spacing is 0.33m, the plant spacing is 15cm, and each parent has one row. The cultivation management measures in the field test are consistent with the local field management. Seedling pair of parents and 543 F 2 Individual plants were sampled. F 2 The tillering survey was carried out on a single plant, and Microsoft Excel 2010 was used for statistical analysis of the data.
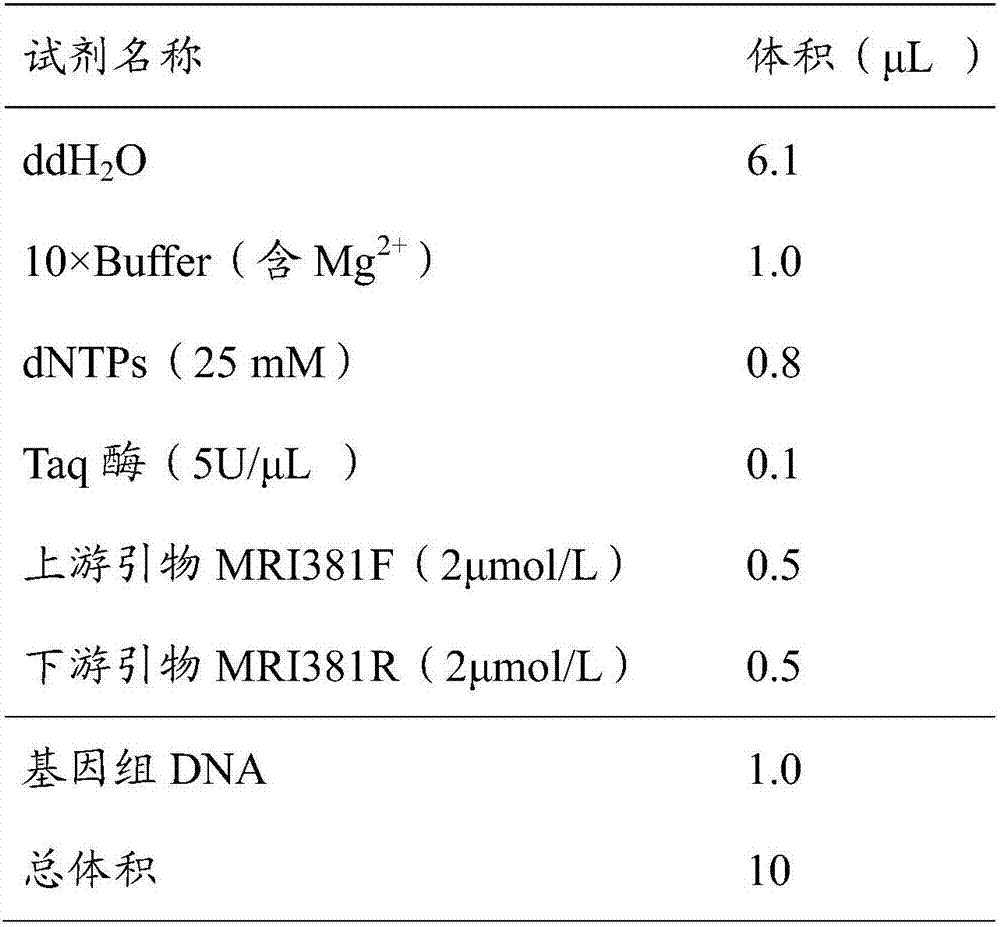
[0036] Using the RAD-seq method, the parental and 543 F 2 The genomic DNA of a single plant is tested for DNA quality, the quali...
Embodiment 2
[0048] Embodiment 2: millet F 2 Population library sequencing
[0049] The millet F obtained in embodiment 1 2 The group database construction method is as follows:
[0050] (1) Weigh 1.0g of fresh leaves, cut them into pieces and put them in a mortar, grind them with liquid nitrogen, add 3mL 1.5×CTAB, grind them into a homogenate and transfer them to a 15mL centrifuge tube, then add 1mL 1.5×CTAB into the mortar Rinse and transfer to a centrifuge tube. After mixing, place in a water bath at 65°C for 30 minutes, and shake slowly from time to time during this period.
[0051] The 1.5×CTAB formula is as follows (1L): CTAB 15g, 1mol / L Tris.Cl (pH 8.0) 75mL, 0.5mol / L EDTA 30mL, NaCl 61.4g, add deionized water to 1L, before use Mercaptoethanol was added to a final concentration of 0.2% (2 ml).
[0052] (2) After cooling to room temperature, add an equal volume of chloroform / isoamyl alcohol (24:1, v / v) and mix gently until the lower layer turns dark green.
[0053] (3) Centrifu...
Embodiment 3
[0074] Example 3: Extraction of Genomic DNA
[0075] For the millet F obtained in Example 1 2 Group, use CTAB method to extract parents and F 2 Genomic DNA of individual plants in the population, the specific method is as follows:
[0076] (1) Weigh 1.0g of fresh leaves, cut them into pieces and put them in a mortar, grind them with liquid nitrogen, add 3mL 1.5×CTAB, grind them into a homogenate and transfer them to a 15mL centrifuge tube, then add 1mL 1.5×CTAB into the mortar Rinse and transfer to a centrifuge tube. After mixing, place in a water bath at 65°C for 30 minutes, and shake slowly from time to time during this period.
[0077] The 1.5×CTAB formula is as follows (1L): CTAB 15g, 1mol / L Tris.Cl (pH 8.0) 75mL, 0.5mol / L EDTA 30mL, NaCl 61.4g, add deionized water to 1L, before use Mercaptoethanol was added to a final concentration of 0.2% (2 ml).
[0078] (2) After cooling to room temperature, add an equal volume of chloroform / isoamyl alcohol (24:1, v / v) and mix gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com