SNP markers for identification of low stone cell content in pear pulp based on high-resolution melting curves and their application
A high-resolution, melting curve technology, applied in the field of SNP markers, can solve the problems that the development and application of SNP molecular markers have not yet been reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] The mensuration of embodiment 1 stone cell content
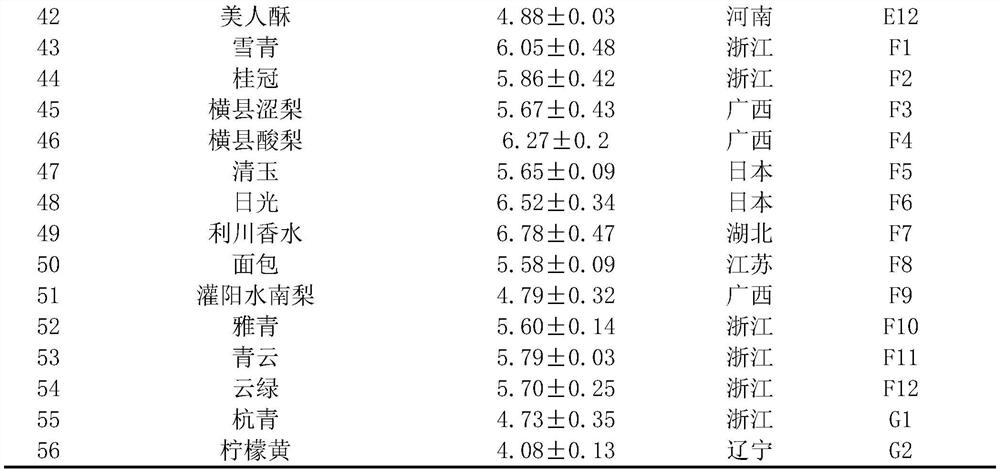
[0025] Stone cells were separated and weighed according to the method of Tao et al. (2009) [6]. Peel and remove the core of the fruit, take 100g of pulp and add distilled water to beat, then dilute the homogenate with distilled water, stir and let it stand at room temperature for 30 minutes, discard the floating matter, resuspend the sediment in 0.5mol / L hydrochloric acid for 30 minutes, discard Floats were washed with distilled water. Repeat the above steps until the stone cells are separated from other cell fragments, and weigh after drying. Table 1 shows the content of stone cells in the flesh of 56 pear natural populations at 35 days after flowering.
[0026] Table 1 Stone cell content in the pulp of 56 pear natural populations at 35 days after flowering
[0027]
[0028]
[0029] Note: Serial numbers 1-30 are varieties with high stone cell content, and 31-56 are varieties with low stone cell content.
Embodiment 2
[0030] Example 2 Development of HRM-specific primers using SNP marker sites Psc397-7 and Psc397-12
[0031] The DNA sequences of SNP markers Psc397-7 and Psc397-12 on the fifth chromosome were searched from the whole genome database of Dangshan Suli, and the 300 bp sequences before and after the site were selected, and the primers for SNP markers were developed and designed according to the principle of primer design (Table 2). Using the designed SNP marker primers to carry out PCR amplification on the genomic DNA of Dangshansu pear, the primers were amplified normally, and the PCR product was in line with the predicted size.
[0032] Table 2 SNP marker primers
[0033]
Embodiment 3
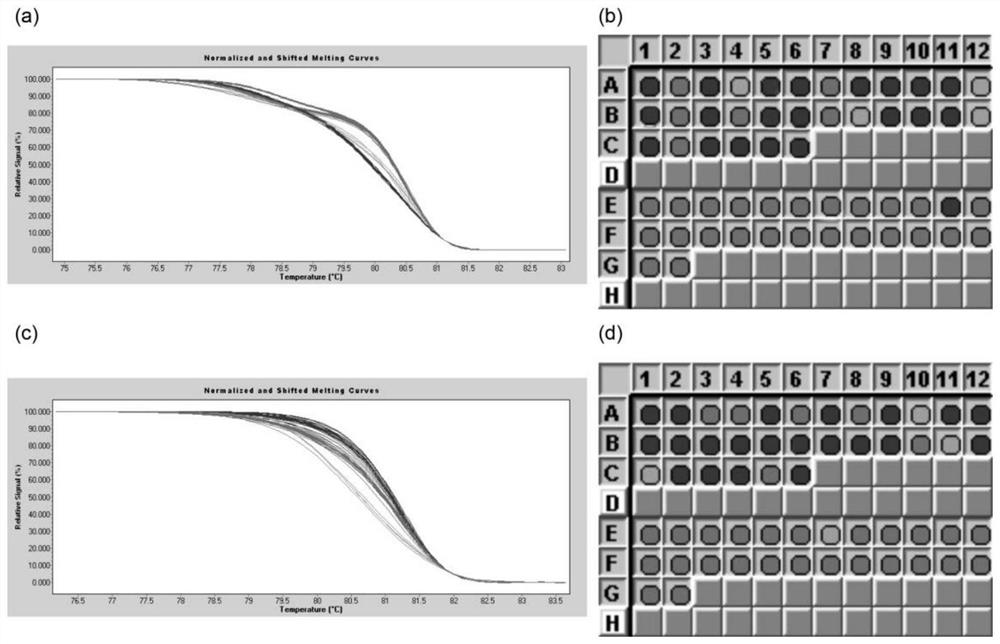
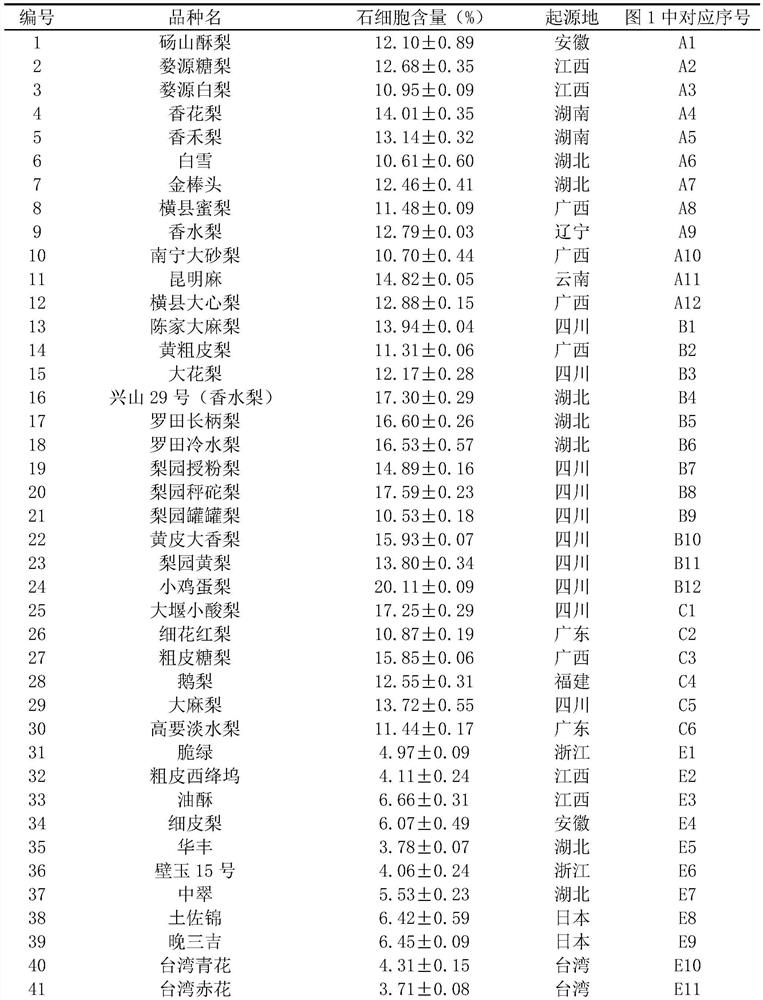
[0034]Example 3 Using HRM (high-resolution melting, high-resolution melting curve) technology to type pear pulp stone cell content
[0035] The HRM reaction system was performed according to the instructions in the LightCycler 480 High Resolution Melting Master kit, and the HRM analysis was performed on a LightCycler480 II fluorescent quantitative PCR instrument. 20μL reaction system: containing 30ng pear genomic DNA template, 1×Master Mix, 2.5mmol / L MgCl 2 , 200 nM of the primers described (Psc397-7-F: SEQ ID NO.1 / Psc397-7-R: SEQ ID NO.2 or Psc397-12-F: SEQ ID NO.3, / Psc397-12-R: SEQ ID NO.4); the amplification program adopts touchdown PCR (touchdown PCR): pre-denaturation at 95°C for 10 minutes, then denaturation at 95°C for 10 seconds, annealing at 65-55°C (0.5°C per cycle) for 10 seconds, and extension at 72°C for 10 seconds The program was performed for 45 cycles. Melting was performed after the PCR cycle was completed. The program was: 95°C for 1 min, 40°C for 1 min, 6...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com