Method and device for detection of gene fusion
A technology of gene fusion and detection method, applied in special data processing applications, instruments, electrical digital data processing, etc., to achieve the effect of low false positives and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0023] According to a typical embodiment of the present invention, the detection method includes S0: repeating sequence filtering, and the repeating sequence filtering step includes: S01, designing a simulated base sequence of 30-75 bp, aligning with the reference genome sequence, finding It is found that the coverage of the simulated base sequence on the reference gene is significantly higher than that of the surrounding sequence, and the region is marked as a highly repetitive region; S02, before performing sequence comparison in step S1 and / or step S42, first The highly repetitive regions in the reference genome are filtered to improve the accuracy of the sequence alignment. Therefore, when the gene sequence to be detected is compared with the reference genome, repetitive sequences can be avoided, so as to improve the efficiency and accuracy of the alignment.
[0024] According to a typical embodiment of the present invention, S41 uses the K-mer algorithm to concatenate to ...
Embodiment 1
[0033] An embodiment of the method for detecting gene fusion of the present invention, wherein:
[0034] Detection object: tumor circulating DNA.
[0035] The concentration of tumor DNA in blood is very low, and most of the sequencing sequences that can be detected by DNA sequencing extracted from blood are derived from tissue DNA sequences. Therefore, in this embodiment, tumor DNA is first identified from tissue DNA, and then gene fusion is identified.
[0036] Detection method: including the following steps:
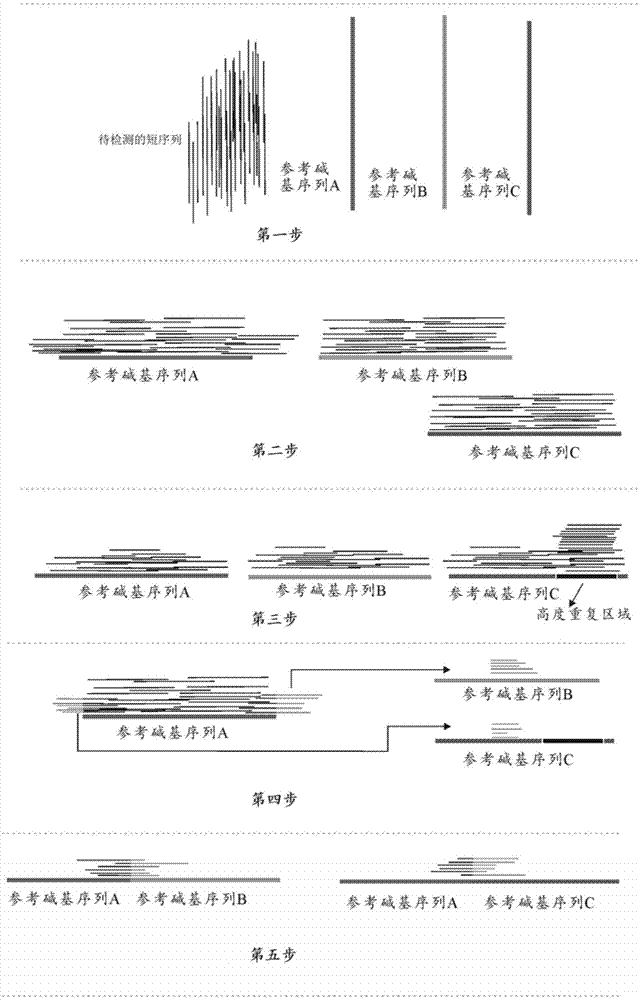
[0037] like figure 1 As shown, the first step: the short lines clustered together on the left are the short sequences to be sequenced; the three lines on the right are the reference sequences of the three genes (named A, B, C, respectively);
[0038] Step 2: Use sequence alignment software to align the short sequence to be sequenced to the reference gene sequence;
[0039] Step 3: Design a 30-75bp simulated base sequence, and use the 30-75bp simulated base sequence...
Embodiment 2
[0048] Three standard products with gene fusion were ordered from the invitrogen website, and after being processed by the gene fusion detection device of the present invention, all the gene fusion sequences were identified, and the positions of the gene fusions were consistent with the results of the standard products; the gene fusion of the standard products The position of the break is fixed (the standard does not give the fusion position), not random, so as long as the detected fusion gene sequence is correct, the position of the gene fusion is also consistent with the standard. The detected frequencies of gene fusions (Table 1 below) were also approximately the same.
[0049] Table 1 Detection effect of the gene fusion detection device of the present invention
[0050] sample name
gene fusion
Standard fusion frequency
Detected fusion frequency
HD664
ALK-EML4
50.00%
43.58%
HD753
ROS1-SLC32A
5.00%
4.20%
HD753...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com