Primer, kit and method for identifying eel germplasm
A technology for identifying eels and eels, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as high cost, need for sequencing, loss of restriction enzyme sites, etc., and achieve practicality Strong performance, easy operation, good stability and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
no. 1 Embodiment
[0034] A primer for identifying eel germplasm, including one or more of the following 4 pairs of primers, the sequences of the 4 pairs of primers are:
[0035] aj S1:TTATGGCTGATTCATCCGAAATT, aj A1 CCTGCTAATGGGTTGAGTACTAAA, fragment size 889;
[0036] r-a S1 TGATGCTGACTTTGCCACTGAT; r-a A2 GTTCGTTCCCTTGAAAACCTTGT, fragment size 341;
[0037] bp-m S5 TCCATACTTCTCATACAAAGACCTAT, bp-m A3CTAGTCAACCTACTAATGGGTTTAAT, fragment size 457;
[0038] m-a S4 CCGCCGTCCCATACGTAGGAG, m-a A4 TATTTTGTTTTTCTAGTCAACCTACTAAT, fragment size 678.
[0039] In the process of applying the primers described in this example to identify eel germplasm, multiple primers are used for amplification in one PCR process, which avoids waste of resources and time caused by multiple amplifications, and can effectively improve efficiency. And the primers will not interact with each other.
no. 2 Embodiment
[0041] A kit for identifying eel germplasm, said kit comprising one or more of the following 4 pairs of primers, said 4 pairs of primer sequences being:
[0042] aj S1:TTATGGCTGATTCATCCGAAATT, aj A1 CCTGCTAATGGGTTGAGTACTAAA, fragment size 889;
[0043] r-a S1 TGATGCTGACTTTGCCACTGAT; r-a A2 GTTCGTTCCCTTGAAAACCTTGT, fragment size 341;
[0044] bp-m S5 TCCATACTTCTCATACAAAGACCTAT, bp-m A3CTAGTCAACCTACTAATGGGTTTAAT, fragment size 457;
[0045] m-a S4 CCGCCGTCCCATACGTAGGAG, m-a A4 TATTTTGTTTTTCTAGTCAACCTACTAAT, fragment size 678.
[0046] In the process of using the kit described in this example to identify eel germplasm, multiple primers are used for amplification in one PCR process, which avoids waste of resources and time caused by multiple amplifications, and can effectively improve efficiency , and the primers will not interact with each other.
no. 3 Embodiment
[0048] A method for identifying eel germplasm, comprising the steps of:
[0049]1. Extract the genomic DNA of any seed or grilled eel product of Japanese eel, American eel, flower eel, Pacific bicolor eel, and European eel;
[0050] 2. Carry out PCR amplification of the eel DNA sample with primers, and the PCR reaction conditions specifically include:
[0051] Pre-denaturation at 94°C for 5 minutes;
[0052] 27 cycles of denaturation at 94°C for 45 seconds, annealing at 58°C for 45 seconds, and extension at 72°C for 45 seconds;
[0053] Extension at 72°C for 5 minutes.
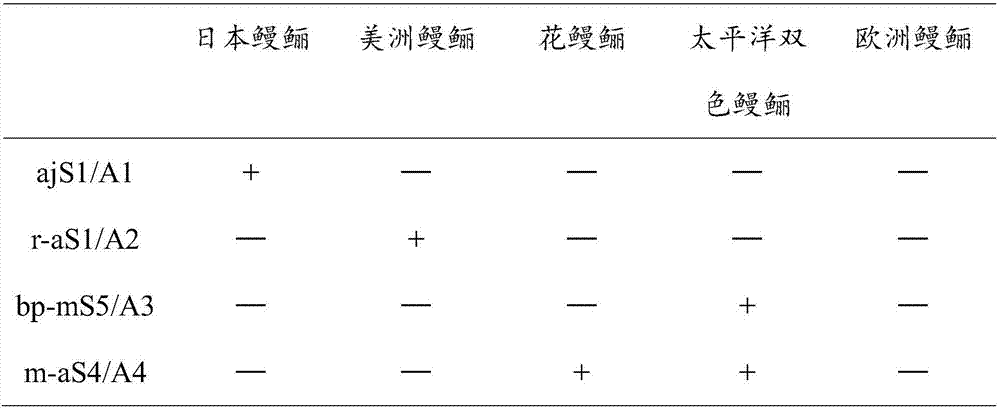
[0054] 3. Using agarose gel electrophoresis to detect the negative and positive bands and the size can quickly identify eel germplasm, the results are as follows:
[0055] Table PCR results of different primers
[0056]
[0057] Note: + means positive, - means negative
[0058] The size of fragments amplified by different primers of this method is different. The Japanese eel is 889bp, the South America...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com