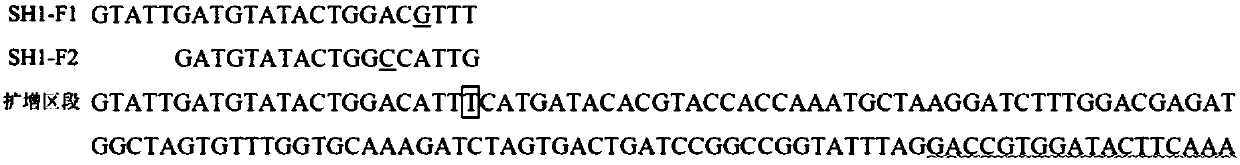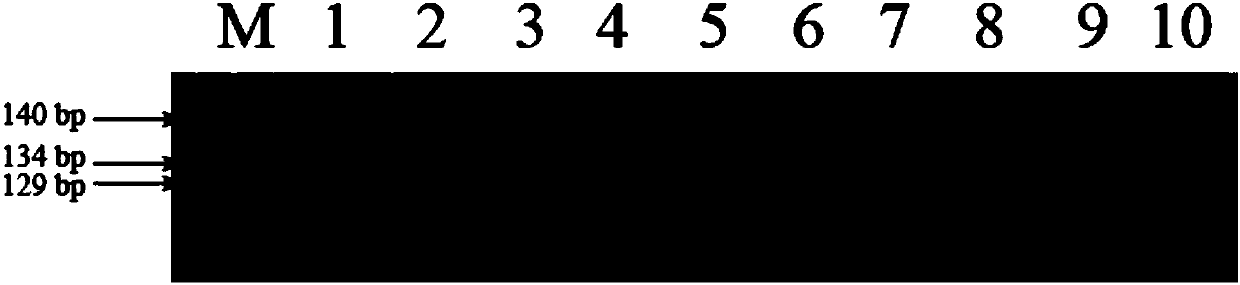Functional marker of rice seed shattering gene qSH1, and application of functional marker
A shattering, SH1-F2 technology, applied in the field of agricultural biology, can solve the problems of difficult shattering, differences in expression parts, affecting the breeding process, etc., and achieve the effects of speeding up the breeding process, improving the breeding efficiency, and improving the breeding efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
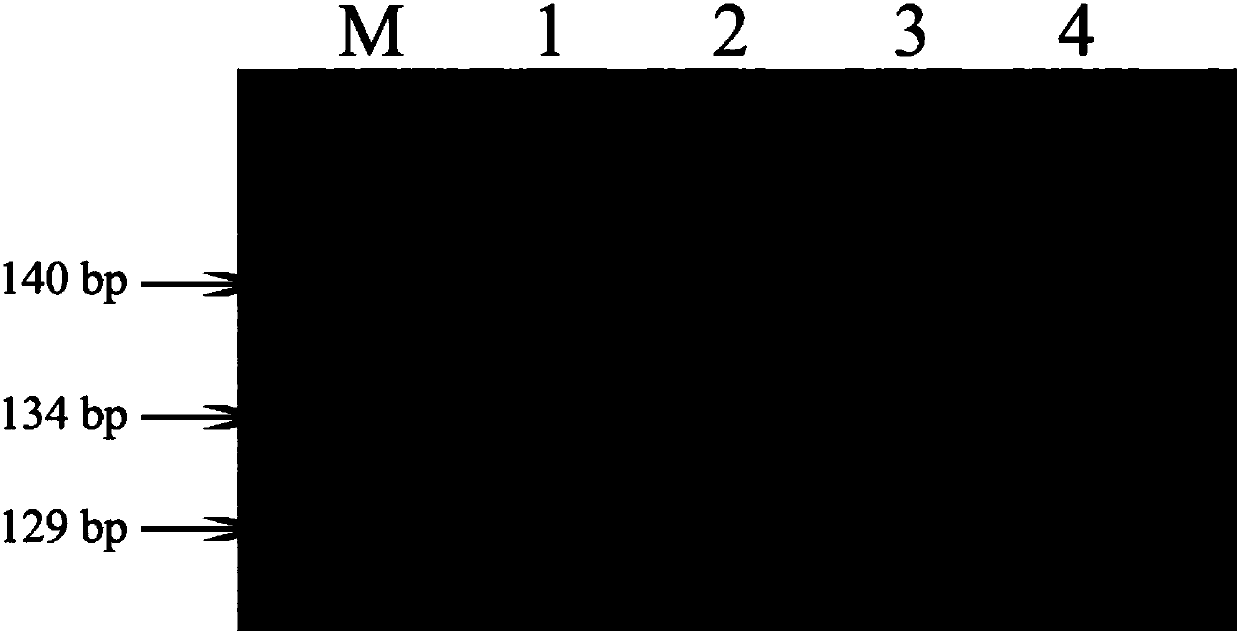
[0037] Example 1 Detection of 4 Rice Varieties Using Functional Marker SH1
[0038] (1) Genomic DNA was extracted from 4 rice varieties (Nipponbare, which is not easy to shatter, and Dongxiang wild rice, Zengcheng wild rice and Gaozhou wild rice, which are easy to shatter): rice leaves were taken respectively, and rice genomic DNA was obtained by TPS simple method.
[0039] (2) PCR amplification
[0040] The PCR reaction system is a 20 μl reaction system: 2.0 μl of 10×PCR buffer; 0.5 μl of 10 mM dNTPs; 0.5 μl of 10 μM three primers (SH1-F1, SH1-F2 and SH1-R); 0.2 μl of Taq DNA polymerization Enzyme, 2.0 μl of template DNA; 13.8 μl of ddH2O.
[0041] The PCR reaction program was: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 45 sec, annealing at 48°C for 30 sec, extension at 72°C for 1 min, and 34 cycles; finally, the amplified product was obtained after extension at 72°C for 5 min.
[0042] (3) Detection of amplification products
[0043] The amplified produc...
example 2
[0047] Example 2 Using functional marker SH1 to detect "three-line" rice breeding parents
[0048] (1) Genomic DNA of 332 restorer lines, 84 sterile lines and 81 maintainer lines were extracted respectively: rice leaves were taken respectively, and rice genomic DNA was obtained by TPS simple method.
[0049] (2) PCR amplification
[0050]The PCR reaction system is a 20 μl reaction system: 2.0 μl of 10×PCR buffer; 0.5 μl of 10 mM dNTPs; 0.5 μl of 10 μM three primers (SH1-F1, SH1-F2 and SH1-R); 0.2 μl of Taq DNA polymerization Enzyme, 2.0 μl of template DNA; 13.8 μl of ddH 2 O.
[0051] The PCR reaction program was: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 45 sec, annealing at 48°C for 30 sec, and extension at 72°C for 1 min, 34 cycles; finally, the amplified product was obtained after extension at 72°C for 5 min.
[0052] (3) Detection of amplification products
[0053] The amplified product was electrophoresed in a 6% (w / w) polyacrylamide denaturing gel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com