Maize PILNCR1 and application thereof in regulating and detecting maize tolerance to low-phosphorus stress
A low-phosphorus-resistant, sequence-listed technology that can be used in applications, DNA/RNA fragments, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
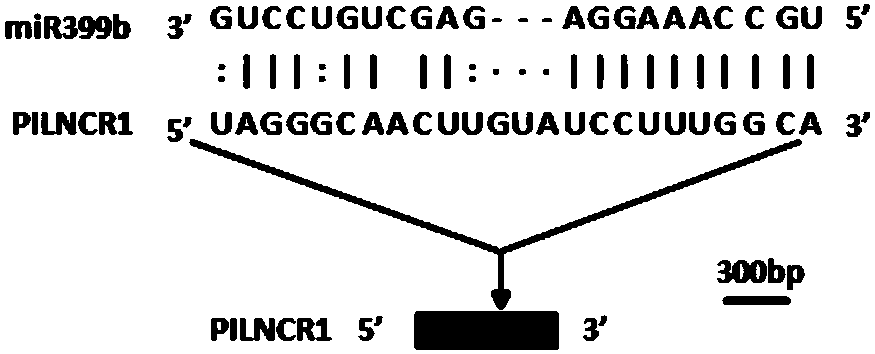
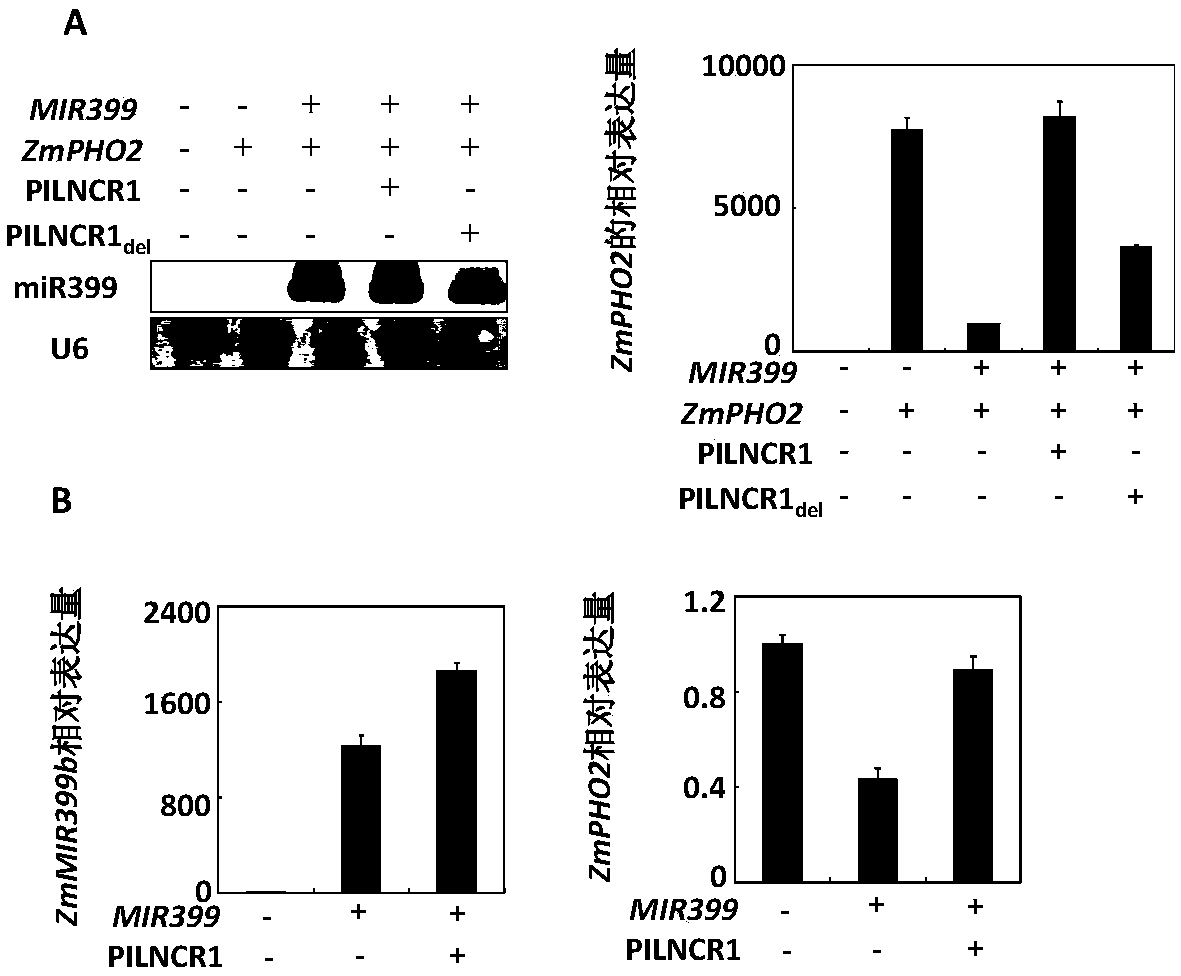
[0054] Example 1. Maize PILNCR1 inhibits the shearing of PHO2 by miR399
[0055] This example provides a LncRNA derived from the low phosphorus sensitive inbred line 31778, its name is PILNCR1, the sequence of PILNCR1 is the 1-649th position of sequence 1 in the sequence table, and the first position of sequence 1 is the 5' of PILNCR1 end. There is a sequence complementary to the mature sequence of miR399 at the 404-428bp of the 5' end of PILNCR1, and there is a 3bp protrusion between the 10th and 11th bases of miR399 in PILNCR1, as shown in figure 1 shown.
[0056] In order to study the effect of PILNCR1 on miR399 and miR399 target gene PHO2 in maize, the inventors combined PILNCR1 full length, PILNCR1 del (remove the sequence complementary to miR399), the full length of miR399 and PHO2 were respectively connected to the pCPB vector, and transformed into protoplasts of tobacco and maize, and the expression of PHO2 was detected. The specific operation method is as follows: ...
Embodiment 2
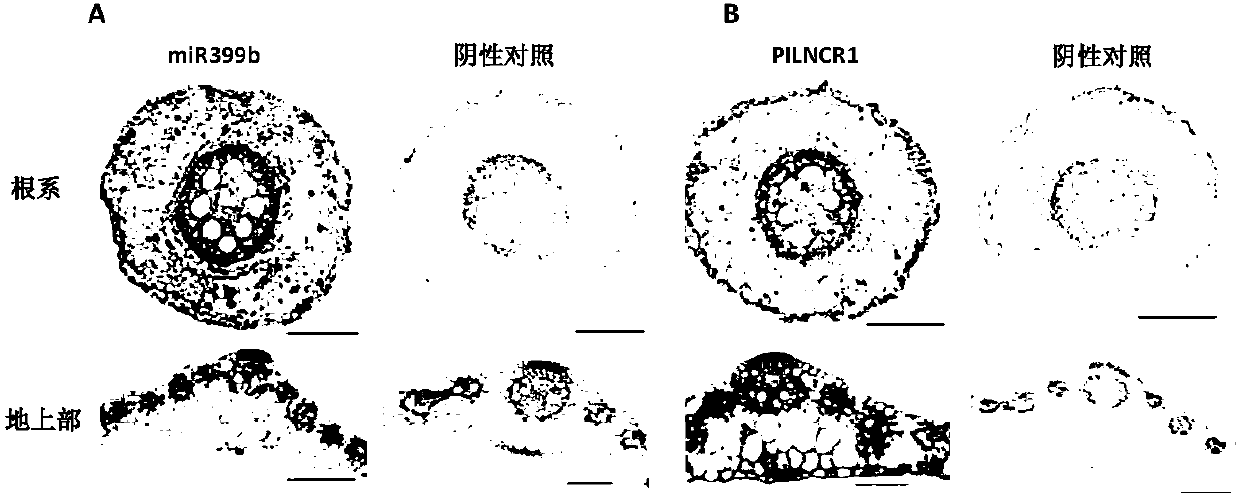
[0090] Example 2, the expression of PILNCR1 in maize inbred lines with different low phosphorus tolerance characteristics
[0091] Determination of low-phosphorus-sensitive maize inbred lines (31778, FR19, 1538, JI419) and low-phosphorus-tolerant maize inbred lines (CCM454, XI14, HAI9-21, Q1261) under normal and low-phosphorus stress conditions by absolute quantitative method The expression of PILNCR1 in .
[0092] Normal treatment (normal phosphorus condition) and low phosphorus treatment (low phosphorus stress condition) methods are as follows:
[0093] Select plump corn seeds of the same size, soak them with 3% NaClO for 20 minutes, and rinse them with sterilized distilled water for 3-4 times. Soak the seeds in distilled water at room temperature for 6 hours, and then bury the seeds in quartz sand (soak the quartz sand with hydrochloric acid overnight, rinse with distilled water 2-3 times); culture at 28°C for 6-7 days, when one leaf and one heart grow (about day), select...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com