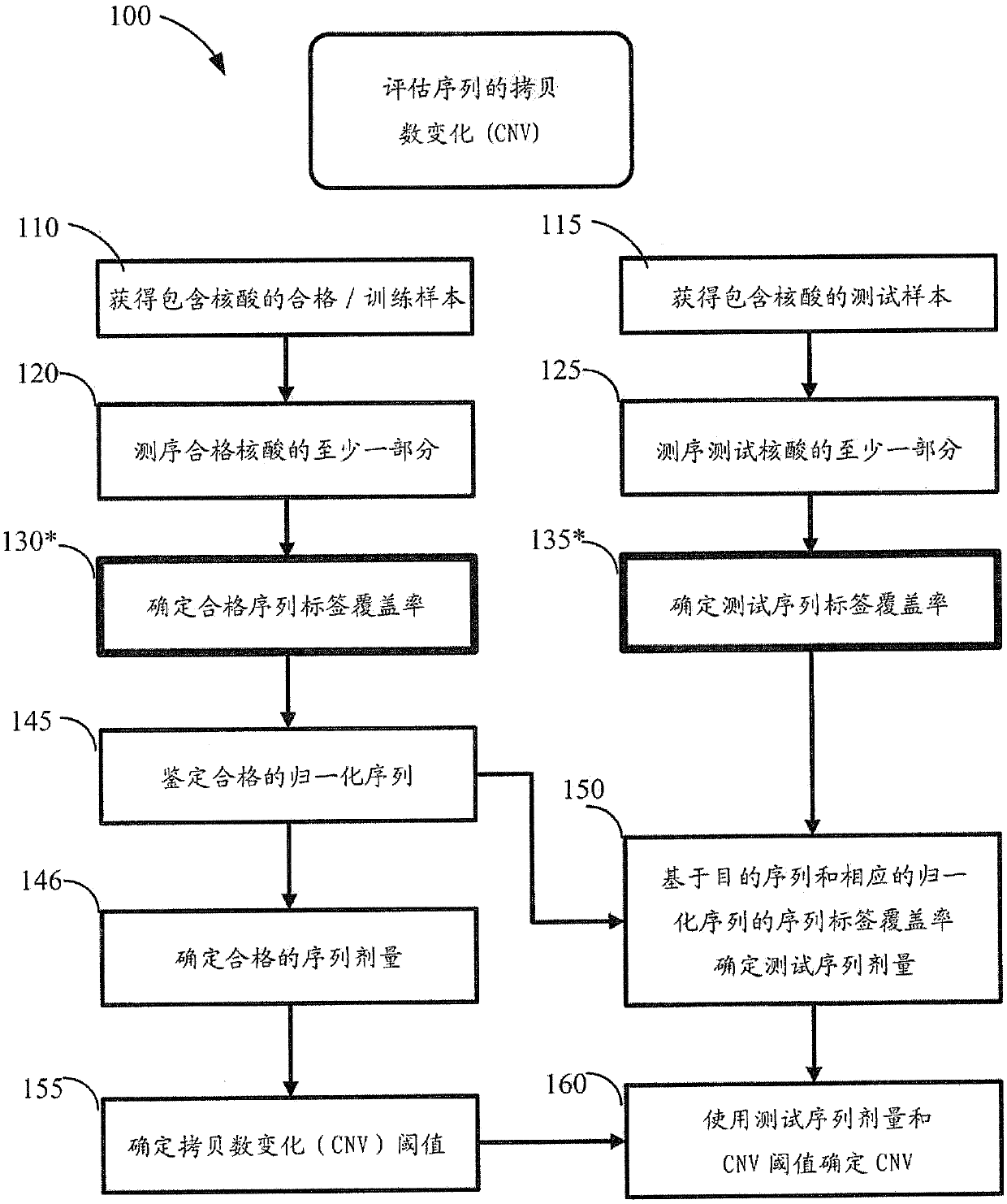
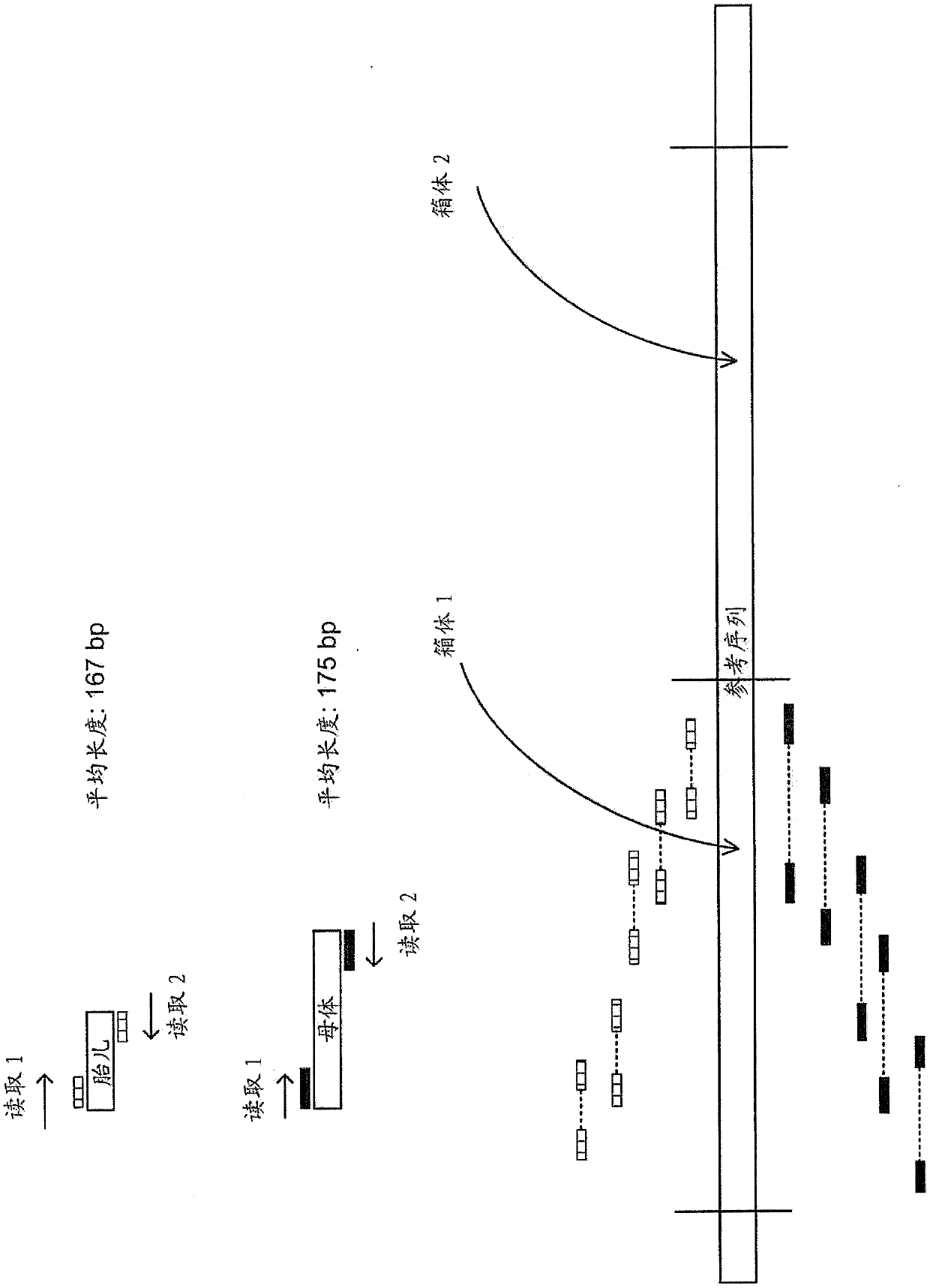
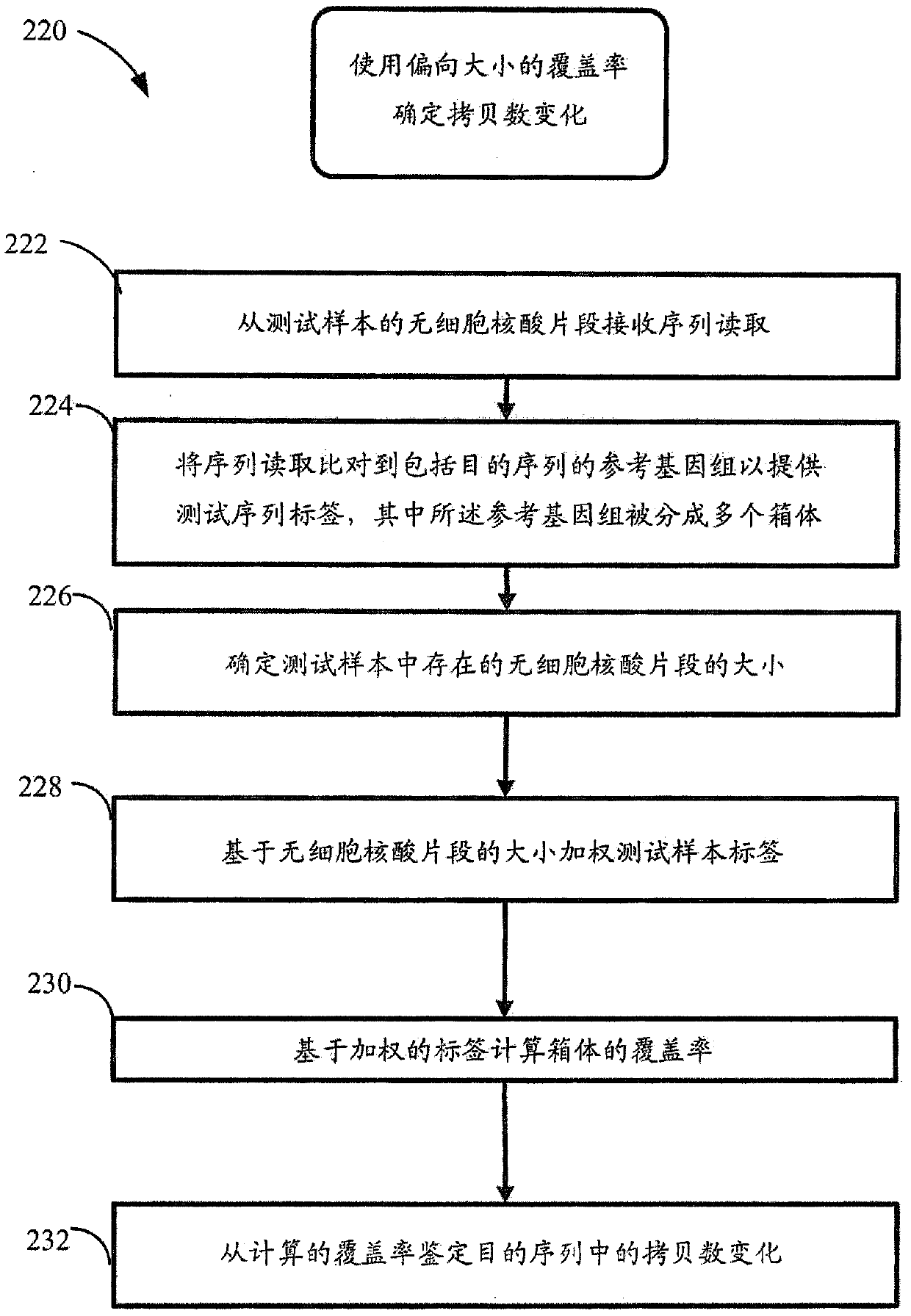
USing cell-free DNA fragment size to determine copy number variations
A copy number, fragment technique used in the field of determining copy number variation using cell-free DNA fragment size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0566] Example 1: Preparation and sequencing of initial and enriched sequencing libraries
[0567] a. Preparation of sequencing library - abbreviated protocol (ABB)
[0568] All sequencing libraries, naive and enriched, were prepared from approximately 2 ng of purified cfDNA extracted from maternal plasma. Use NEBNext TM The reagents of DNA Sample Prep DNA Reagent Set 1 (Part No.E6000L; NewEngland Biolabs, Ipswich, MA) were used for library preparation as follows Because cell-free plasma DNA is fragmented in nature, no further fragmentation by nebulization or sonication was performed on plasma DNA samples. according to End Repair Module by mixing cfDNA with NEBNext in a 1.5ml centrifuge tube TM DNA Sample Prep DNA Reagent Set 1 provided in 5 μl 10X phosphorylation buffer, 2 μl deoxynucleotide solution mix (10 mM each dNTP), 1 μl 1:5 diluted DNA polymerase I, 1 μl T4 DNA polymerase and 1 μl T4 polynuclear Nucleotide kinase incubation for 15 min at 20°C converted ov...
Embodiment 2
[0578] Non-invasive prenatal testing using fragment size
[0579] introduce
[0580] Since its commercial launch in late 2011 and early 2012, noninvasive prenatal testing (NIPT) of cell-free DNA (cfDNA) in maternal plasma has rapidly become the method of choice for screening pregnant women at high risk for fetal aneuploidy. This method is mainly based on the isolation and sequencing of cfDNA from maternal plasma and counting the number of cfDNA fragments aligned to specific regions of the reference human genome (ref: Fan et al, Lo et al). These DNA sequencing and molecular counting methods allow high-precision determination of the relative copy number of each chromosome in the genome. High sensitivity and specificity for the detection of trisomies 21, 18 and 13 have been reproduced in several clinical studies (ref, citing Gil / Nicolaides meta-analysis).
[0581] More recently, additional clinical studies have shown that this approach can be extended to the general obstetr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com