Screening method and device for novel CRISPR-Cas system
A new type of candidate region technology, applied in the field of gene editing, can solve problems such as endogenous RNA interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
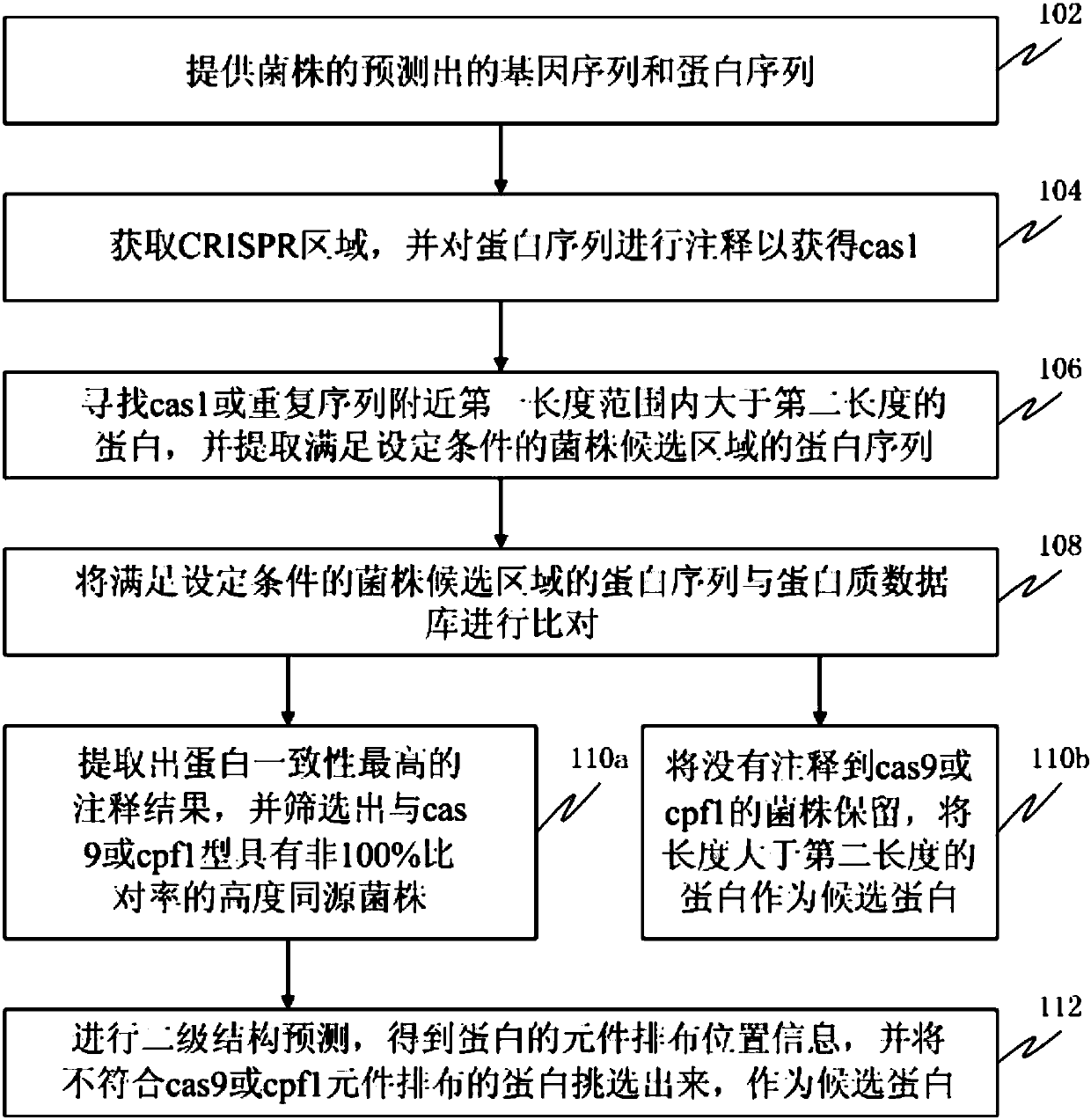
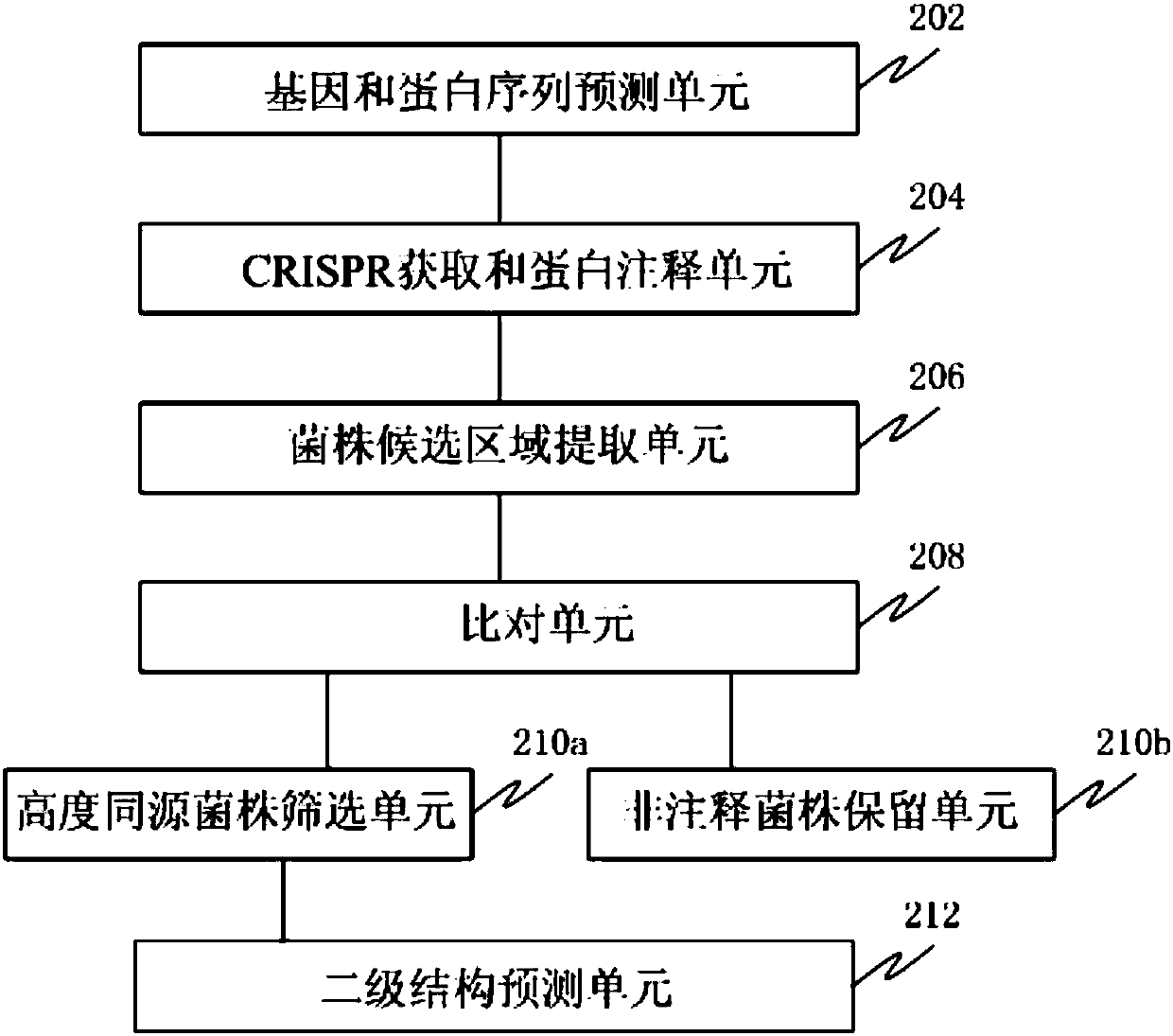
[0071] This example is used to prove that the method of the present invention can effectively reduce the number of candidate strains and candidate proteins. In this embodiment, according to figure 1 The method shown was carried out. Specifically, use MetaGeneMark (version 2.8) software to predict the gene sequence and protein sequence of each bacterial strain; use pilecer (version 1.06) software to find the CRISPR region; use interproscan (version 5.16-55.0) software to perform a Annotation; set the first length to 20kb bases and the second length to 500 amino acids, look for proteins with more than 500 amino acids within 20kb bases near the repeat sequence of cas1 or CRISPR region, and extract the proteins that meet the set conditions The protein sequence of the candidate region of the strain; the set conditions include: (a) there is a repeat sequence in the cas1 and CRISPR region, and it does not belong to type I or type III, and the above-mentioned cas1 and the above-menti...
Embodiment 2
[0074] This example verifies the feasibility and efficiency of the method of the present invention. The experimental conditions and parameters of this embodiment are the same as those of Example 1.
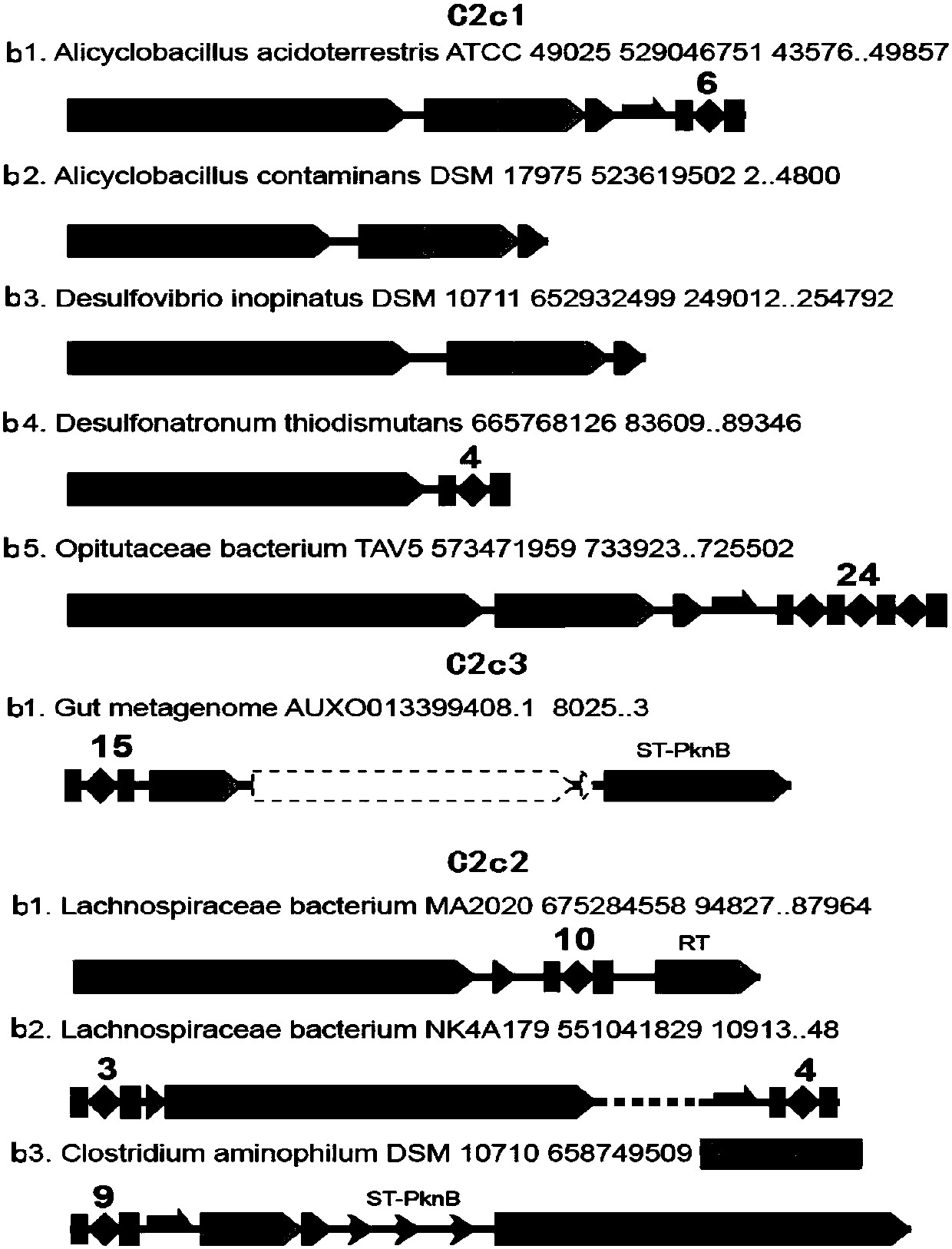
[0075] The screening process of the new CRISPR-Cas system is suitable for analyzing the genome data of a single bacterial species, and selecting the strains that may have a new system, which belongs to the second class (Class2) CRISPR-Cas system other than cas9 and cpf1. In order to verify the feasibility and efficiency of the process, the CRISPR-Cas system, type I system, type III system, and CRISPR-cas9 of c2c1, c2c2, and c2c3 non-cas9 and cpf1 (Class 2) were downloaded from the NCBI database. System, CRISPR-cpf1 system, and genome information of strains with both cas9 system and cpf1 system for process verification. A total of 14 strains were validated to verify the feasibility and efficiency of the screening process for the new CRISPR-Cas system.
[0076] 1) Strain informati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com