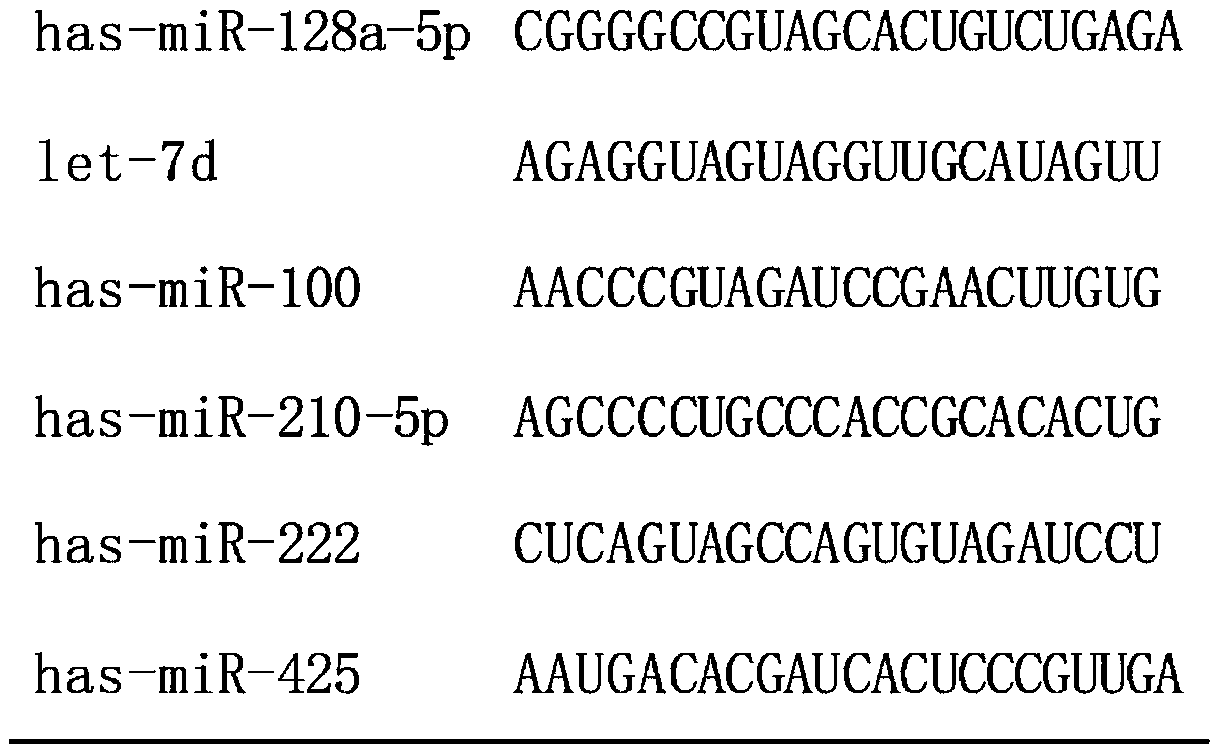Identification of internal reference genes for miRNA detection in osteosarcoma
A technology for osteosarcoma and internal reference, applied in the field of identification of internal reference miRNA genes, to achieve the effects of easy detection, improved sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0024] Example 1 Analysis of miRNA expression in peripheral blood of patients with osteosarcoma
[0025] 1. Sample collection
[0026] Peripheral blood of osteosarcoma patients came from hospitalized patients with osteosarcoma, 30 patients with primary osteosarcoma and 20 healthy controls. The patients with primary osteosarcoma and the healthy control group were required to fast for at least 12 hours. At 7:00-8:00 the next morning at room temperature, 10ml of venous blood was drawn into ethylenediaminetetraacetic acid (EDTA) anticoagulant tubes, and peripheral blood was extracted. For mononuclear cell PBMCs, add 1ml Trizol reagent (Invitrogen), mix well, and store the specimen at -80°C for RNA extraction. All blood samples and pathological results should be authentic and reliable, the study was approved by the ethics committee, and the patients gave informed consent.
[0027] 2. Method
[0028] 2.1. Extraction of total RNA from peripheral blood
[0029] According to the in...
Embodiment 2
[0040] The qRT-PCR verification of miRNA in embodiment 2 serum / plasma
[0041] According to the results of chip hybridization, select miRNA molecules that meet the following criteria for further screening by qRT-PCR technology: a) the expression level in osteosarcoma and the control group is in the top 80; b) it is stably expressed in both groups, and the two groups There was no significant difference between them (p≥0.05). According to the above criteria, six miRNA molecules (including hsa-miR-128a-5p, let-7d, hsa-miR-100, hsa-miR-210-5p, hsa-miR-222, hsa- miR-425), the sequences of the six miRNA molecules are shown in Table 1. In addition, since U6 is often used as an internal reference molecule for tissue miRNA detection, U6 was also included in the screening as a candidate molecule in order to verify whether it can be used as an internal reference in serum. By using qRT-PCR technology to verify the above 6 miRNAs and U6 in another group of subjects (including 10 cases of...
Embodiment 3
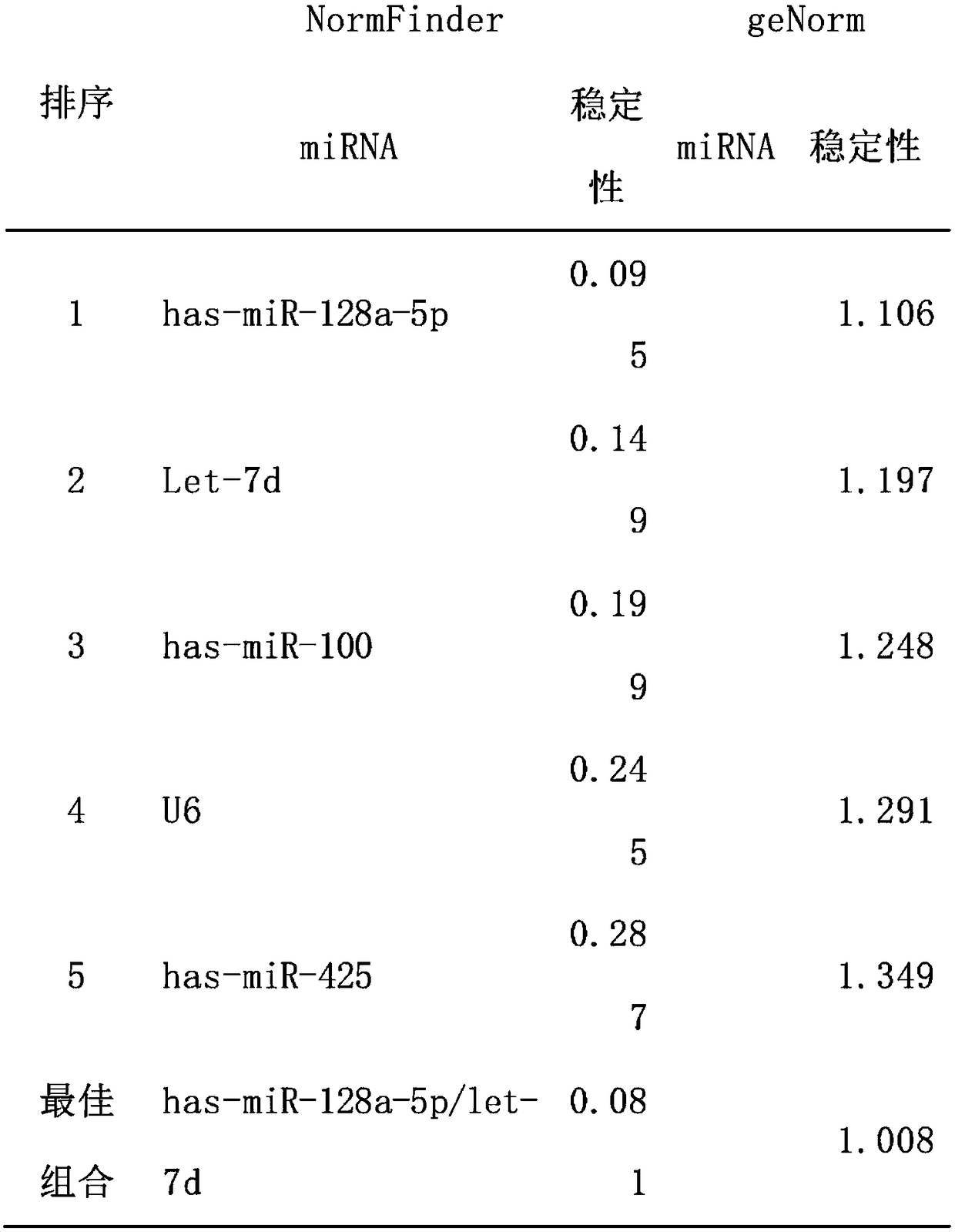
[0058] Example 3 Further verification of expression stability
[0059] According to the results of qRT-PCR screening, the combination of hsa-miR-128a-5p or hsa-miR-128a-5p / let-7d was a candidate internal reference gene with stable expression. The qRT-PCR method was used to verify its stability in a group of new test samples (including 10 osteosarcoma and 10 control samples), and the results showed that it was stably expressed in both osteosarcoma and normal samples, with relatively good stability.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com