Identification of reference gene for detecting lung cancer miRNA
A lung cancer and internal reference technology, applied in the field of internal reference miRNA gene identification, to achieve the effect of improving sensitivity and specificity, and easy detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0024] Example 1 Analysis of miRNA expression in peripheral blood of patients with lung cancer
[0025] 1. Materials and methods
[0026] 1. Materials
[0027] Peripheral blood of lung cancer patients was obtained from hospitalized cases of lung cancer. Peripheral blood was collected from 30 lung cancer patients and 20 normal people.
[0028] 2. Method
[0029] 2.1. Extraction of total RNA from peripheral blood
[0030] According to the instructions of the Triazol kit, total RNA was extracted from the peripheral blood cells of lung cancer patients and normal people, the integrity of the RNA was verified by gel electrophoresis, and the concentration and purity of the RNA were determined with a nucleic acid protein analyzer. Proceed as follows:
[0031] (1) Collect peripheral blood from the patient, collect cells by centrifugation, add 1 mL of Trizol reagent to each tube, mix well, and place at room temperature for 5 minutes. Add 200 μL of chloroform, shake vigorously for 1...
Embodiment 2
[0041] The qRT-PCR verification of miRNA in embodiment 2 serum / plasma
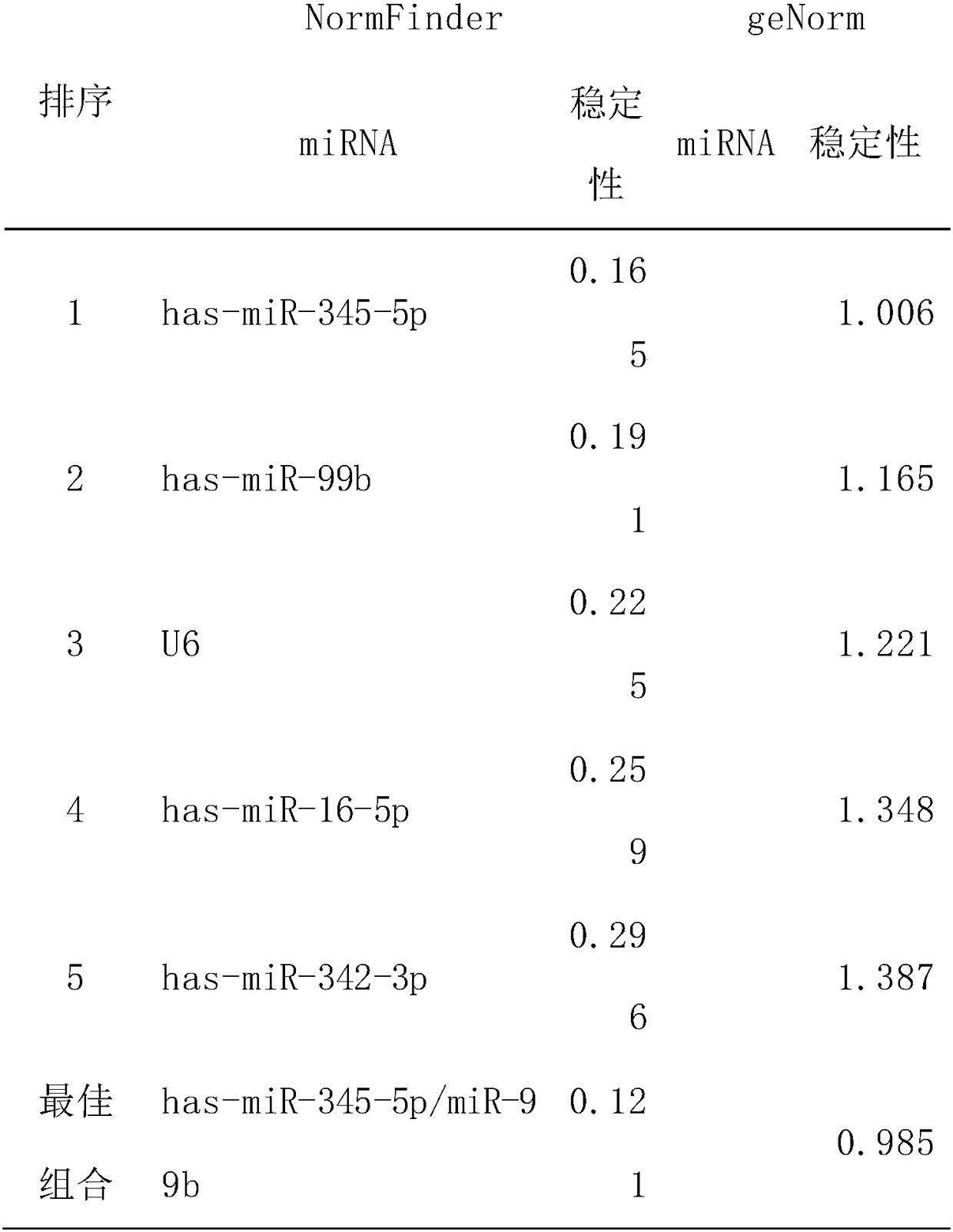
[0042]According to the results of chip hybridization, miRNA molecules meeting the following criteria were selected for further screening by qRT-PCR technology: a) the expression levels in lung cancer and the control group were in the top 100; b) they were stably expressed in both groups, and There was no significant difference between them (p≥0.05). According to the above criteria, a total of 9 qualified miRNA molecules were selected (including hsa-miR-140-5p, hsa-miR-26a, hsa-miR-99b, hsa-miR-106b, hsa-miR-151-5p, hsa-miR-23a, hsa-miR-342-3p, hsa-miR-345-5p, hsa-miR-16-5p), the sequences of nine miRNA molecules are shown in Table 1. In addition, since U6 is often used as an internal reference molecule for tissue miRNA detection, U6 was also included in the screening as a candidate molecule in order to verify whether it can be used as an internal reference in serum. By using qRT-PCR technology to verify ...
Embodiment 3
[0059] Example 3 Further verification of expression stability
[0060] According to the results of qRT-PCR screening, has-miR-345-5p or the combination of has-miR-345-5p and has-miR-99b are candidate internal reference genes with stable expression. The qRT-PCR method was used to verify its stability in a new set of tested samples (including 12 liver cancer and 12 control samples), and the results showed that it was stably expressed in both liver cancer and normal samples, and had a good stability.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com