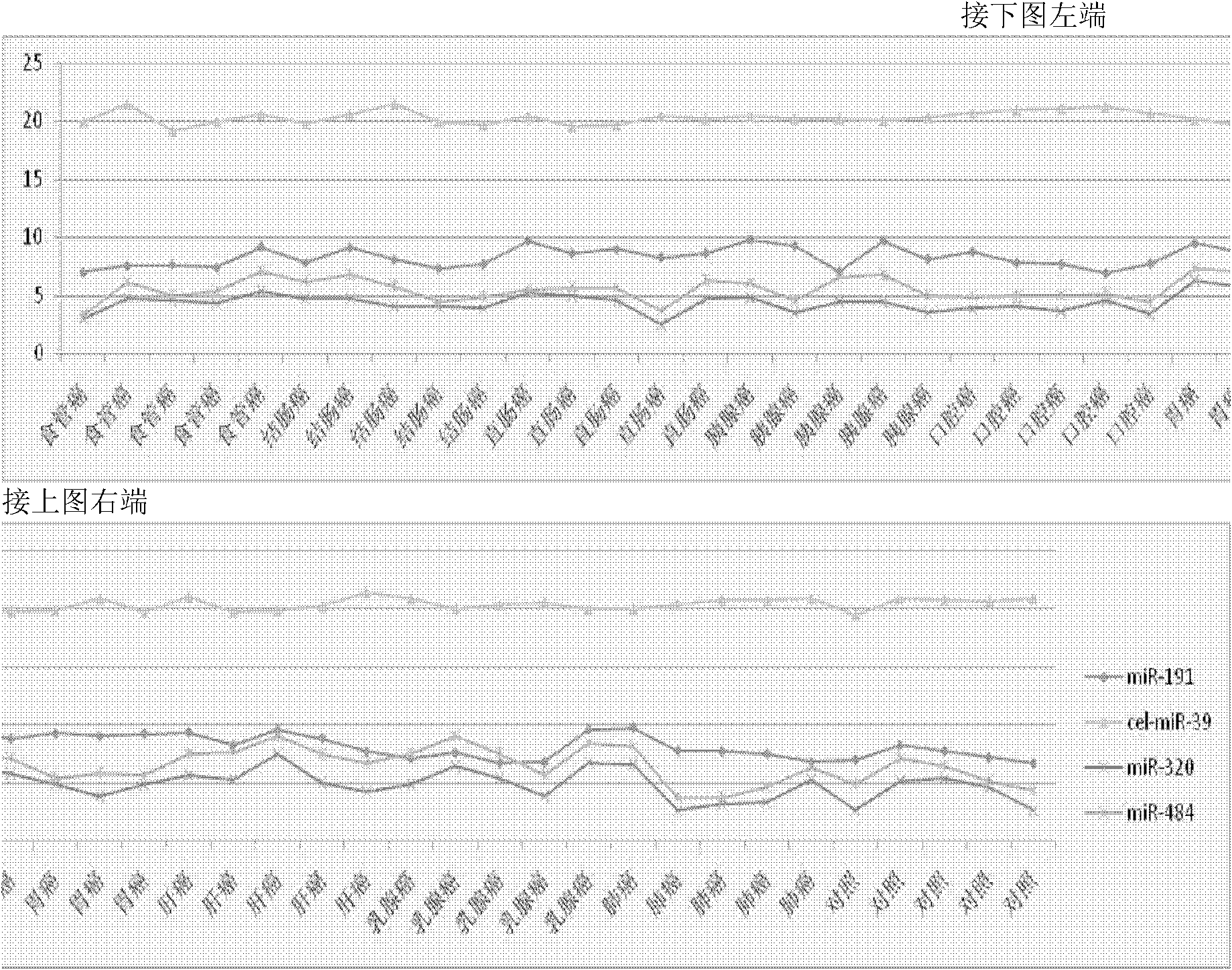Internal reference for detecting miRNA (micro Ribonucleic Acid) in serum/blood plasma and application of internal reference
An internal reference and plasma technology, applied in the fields of genetic engineering and oncology, can solve the problems of uncontrollable biological differences and restrictions on clinical transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
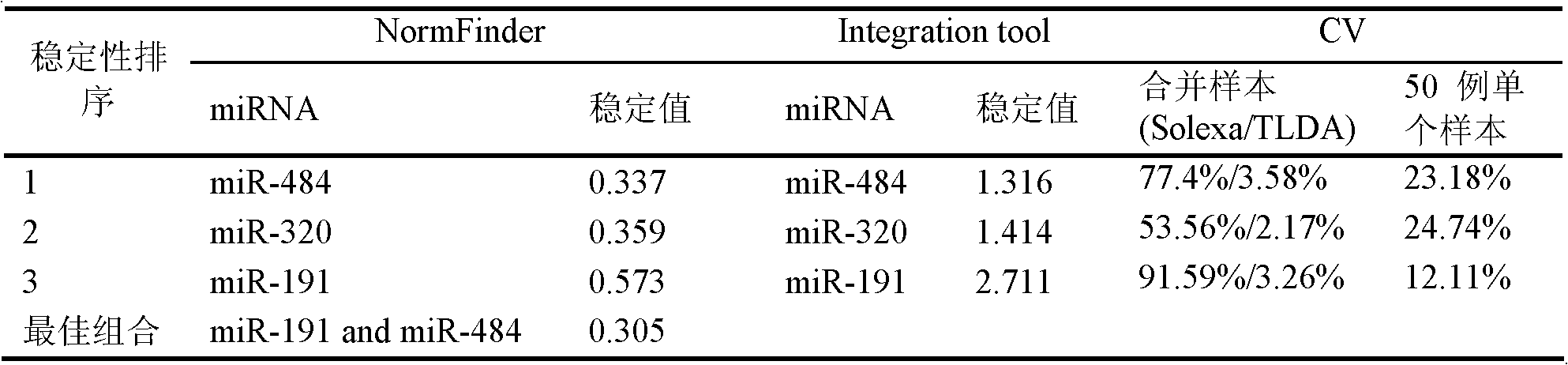
[0069] The collection of embodiment 1 sample and the arrangement of sample data
[0070] The inventor has collected a large amount of serum / plasma samples of various tumor patients and healthy controls from multiple tertiary first-class hospitals in Jiangsu Province since July 2003, and the cases and control samples used for research are collected at the same period, and sampling, analysis The installation and storage conditions were uniform, and by sorting out the sample data, the inventor selected 354 samples that met the following criteria as experimental samples for Solexa sequencing, TLDA chip detection, and a series of subsequent qRT-PCR verifications:
[0071] 1. Cancer patients diagnosed by pathology, including cases of lung cancer, breast cancer, gastric cancer, liver cancer and cervical cancer in the internal reference screening stage, and esophageal cancer, colon cancer, rectal cancer, pancreatic cancer, oral cancer, gastric cancer in the internal reference verificat...
Embodiment 2
[0075] Example 2 Solexa sequencing experiment of miRNA in serum / plasma
[0076] The above qualified samples include 60 cases of lung cancer (30 cases of long survival group and 30 cases of short survival group), 48 cases of breast cancer (24 cases / group), 40 cases of gastric cancer (20 cases / group), 30 cases of liver cancer, 30 cases of cervical cancer and 48 healthy male controls, 48 healthy female controls. These 10 groups of people were subjected to the Solexa sequencing test (the kit was purchased from ABI Company) to obtain relevant results. The specific steps are:
[0077] 1. Take 50ml of serum / plasma from each group of samples, and add an equal volume of Trizol reagent;
[0078] 2. Phase separation: place at room temperature for 15 minutes, then add chloroform according to the volume ratio of 0.2ml chloroform / 1ml Trizol reagent, shake for 15s, room temperature for 15 minutes, centrifuge at 12,000g, 4°C for 15 minutes;
[0079] 3. Transfer the aqueous phase to a new...
Embodiment 3
[0089] TLDA chip detection of miRNA in embodiment 3 serum / plasma
[0090] The above 10 groups of samples subjected to Solexa sequencing were detected by TLDA chip (the kit was purchased from ABI Company) to obtain relevant results. The specific steps are:
[0091] 1. Take 600 μl of serum / plasma from each group and add 3 times the volume of Trizol reagent;
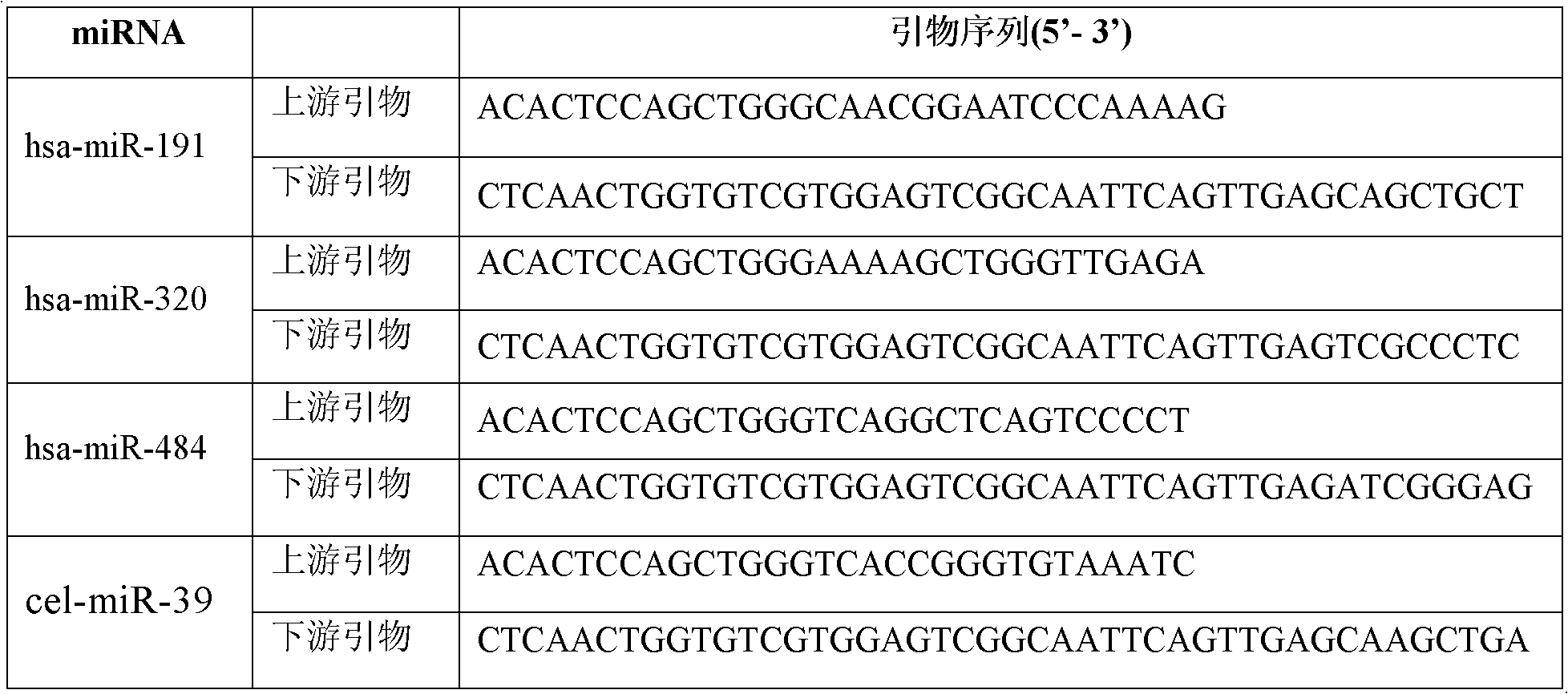
[0092] 2. Phase separation: place at room temperature for 15 minutes, add cel-miR-39 (TAKARA) with a final concentration of 10-4pmol / μl as an internal reference, then add chloroform equal to the volume of serum / plasma, shake for 50s, room temperature for 15 minutes, 14,000rpm, Centrifuge at 4°C for 15 minutes;
[0093] 3. RNA precipitation: transfer the water phase to a new 15ml centrifuge tube, add 1.5 times the volume of the water phase in absolute ethanol, and mix well;
[0094] 4. Enrich RNA with QIAGEN miRNeasy kit: pipette 700 μl of sample into the spin column each time, centrifuge at 14,000 rpm for 15 s, discard t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com