A method and system for detecting epistasis in complex diseases based on chromatin regulatory loops
A loop detection and chromatin technology, applied in hybridization, genomics, proteomics, etc., can solve the problems of large computing resources and time, consumption, and epistasis that has not been seen, so as to reduce the amount of calculation and reduce false negatives The effect of the result
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] The present invention will be described in further detail below in conjunction with the accompanying drawings and embodiments. The examples are only used to explain the content of the present invention.
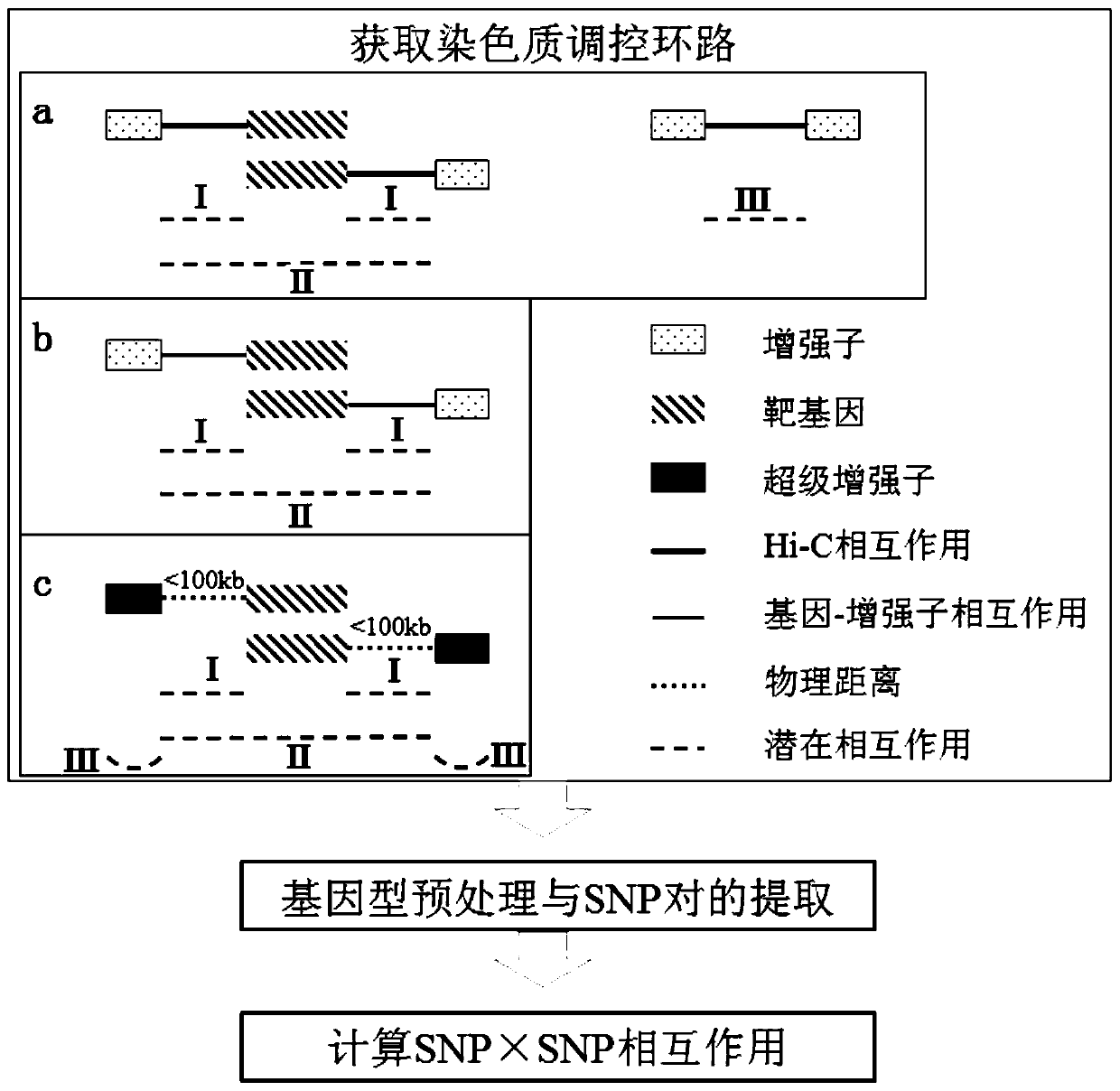
[0032] Based on the existing chromatin long-range interaction data, gene-enhancer regulatory data and super-enhancer data, the applicant defined the chromatin regulatory loop that interacts with the target gene, and invented a chromatin-based regulatory loop Methods for detecting epistasis in complex diseases. The basic steps are as follows:
[0033] P1: Collect and organize chromatin long-range interaction data and corresponding chromatin segmentation state data, gene-enhancer regulation data, and super-enhancer data of complex disease-related cell lines;
[0034] P2: Use the above data to establish chromatin regulatory loops that interact with target genes;
[0035] P3: Using the established chromatin regulatory circuit, calculate the SNP interactions in the regul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com