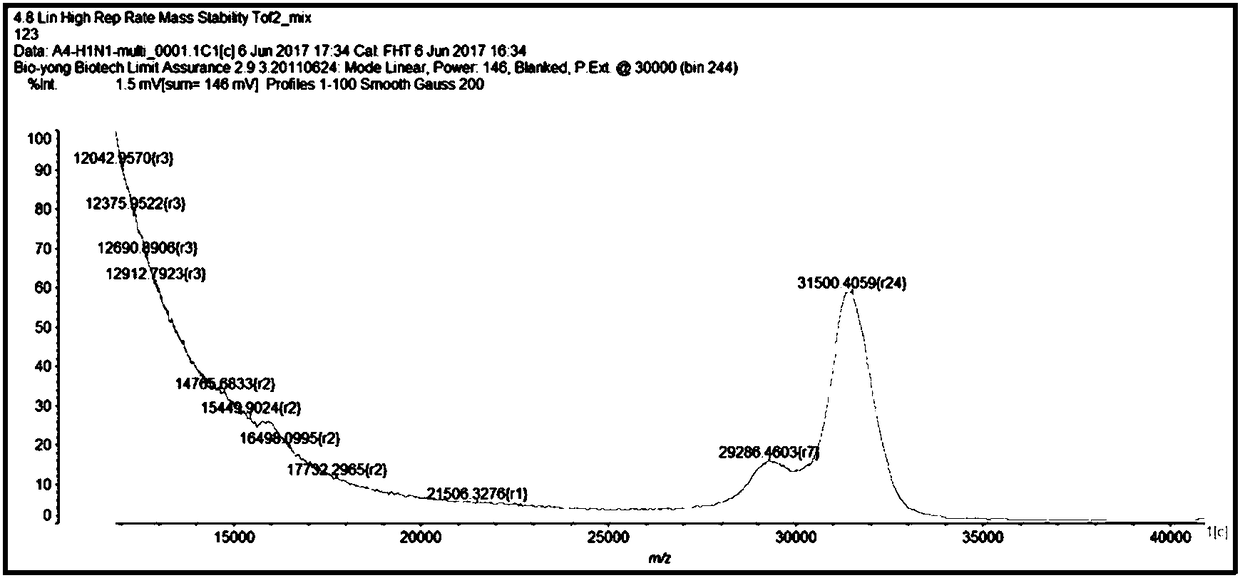
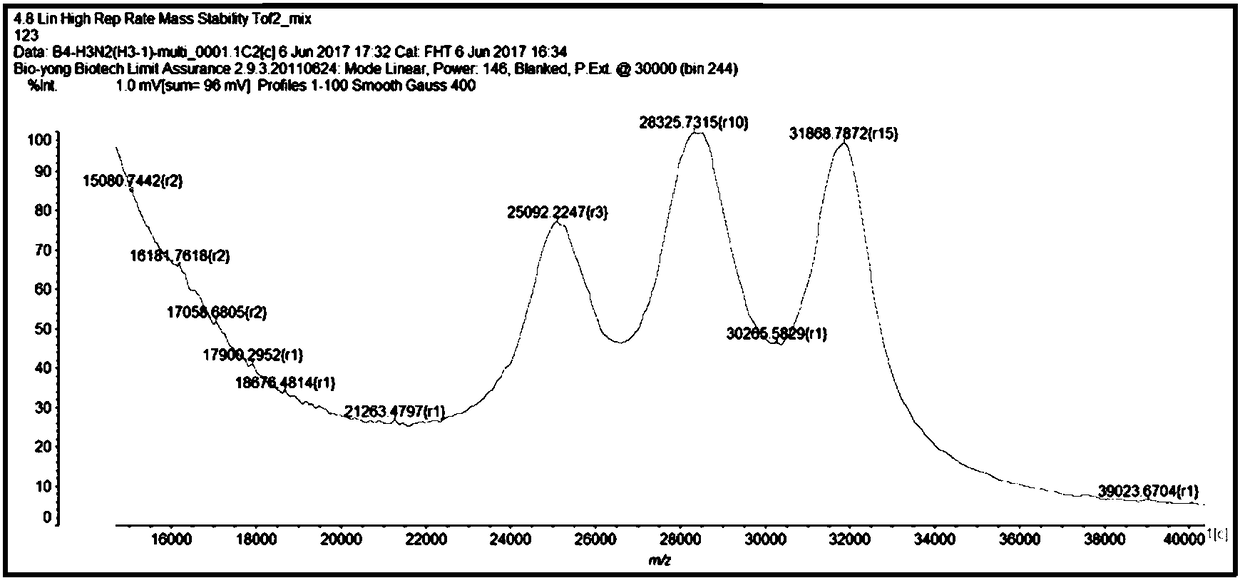
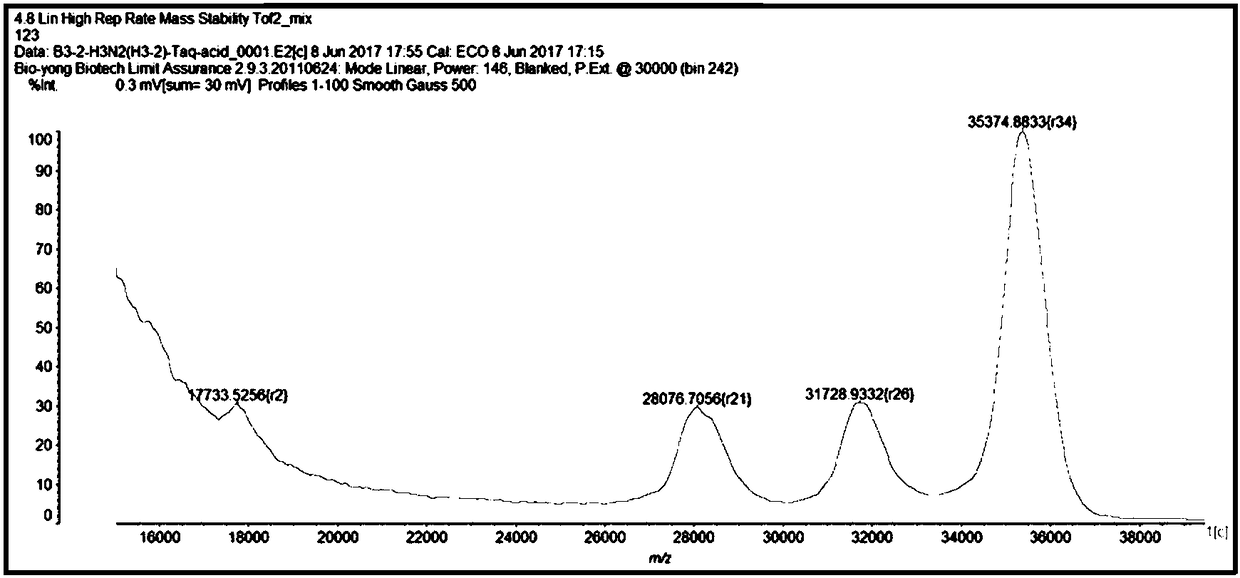
Mass spectrum method for detecting H3N2 fragment multiple PCR product and its product
A mass spectrometry, multiplex technology, applied in the direction of biochemical equipment and methods, DNA / RNA fragments, microbial determination / inspection, etc., can solve the problem of no specific target of influenza virus reported, no reporting scheme, difficult to teach and detect influenza virus, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Example 1. Primer Design
[0088] In order to detect influenza A virus, the following nine nucleic acid sequences of four common subtypes of influenza A virus, H1N1, H3N2, H5N1 and H7N9, were collected. Extract viral RNA, design reverse transcription primers for the target detection segments (M, H1, H3, H5, H7, N1, N2, N9, etc.), reverse transcribe these segments into cDNA, and connect them to the plasmid vector PmdTM19- Transformation was carried out on T SimpleVector, and 9 plasmids respectively containing the 9 nucleic acid sequences were synthesized.
[0089] After identification, the plasmid DNA was extracted, and the concentration of the plasmid DNA was measured using a NanoDrop ND-2000 nucleic acid detector, and the copy number of the DNA was determined as a sensitivity standard to quantify the mother liquor.
[0090] In the following sequences, the sequences corresponding to the selected primers of the present invention are underlined.
[0091] (1) H1 fragment...
Embodiment 2
[0141] 2-fold PCR amplification of embodiment 2.H1N1 subtype influenza A virus
[0142] 1. Synthesize the plasmids of the H1 and M fragments of the H1N1 subtype influenza A virus, and design specific primers corresponding to their conserved sequences. The plasmids and specific primers are all synthesized by Shanghai Jierui Bioengineering Co., Ltd. The plasmid was made into a 10ng / μL working solution, and the primer was made into a 10μM working solution. As shown in Table 1, the following primer pairs were used.
[0143] First primer pair H1:
[0144] Upstream primers:
[0145] SEQ ID NO.1: 5'-TGCTGGATCTGGTATTATC-3',
[0146] Downstream primers:
[0147] SEQ ID NO.2: 5'-TGGGAGGCTGGTGTTTATAG-3';
[0148] Second primer pair M:
[0149] Upstream primers:
[0150] SEQ ID NO.3: 5'-GGCGTTTTGAACAAACCGTC-3',
[0151] Downstream primers:
[0152] SEQ ID NO. 4: 5'-CAATCCTGTCACCTCTGACT-3'.
[0153] 2. PCR amplification
[0154] 1) The composition of the PCR reaction system is a...
Embodiment 3
[0164] Triple PCR amplification of embodiment 3.H3N2
[0165] 1. Synthesize the plasmids of the H3, N2 and M segments of the H3N2 subtype influenza A virus, and design specific primers corresponding to their conserved sequences. The plasmids and specific primers are all synthesized by Shanghai Jierui Bioengineering Co., Ltd. The plasmid was made into a 10ng / μL working solution, and the primer was made into a 10μM working solution. As shown in Table 1, the following primer pairs were used.
[0166] First primer pair H3-1:
[0167] Upstream primers:
[0168] SEQ ID NO.5: 5'-CCGGATGAGGCAACTAGTGA-3',
[0169] Downstream primers:
[0170] SEQ ID NO.6: 5'-GCAGCAAAGCCTACAGCAAC-3';
[0171] Second primer pair N2:
[0172] Upstream primers:
[0173] SEQ ID NO.9: 5'-TATCATCCCCAGTGACACAG-3',
[0174] Downstream primers:
[0175] SEQ ID NO.10: 5'-TGGGAACCAAAACAAGTGTGC-3';
[0176] Third primer pair M:
[0177] Upstream primers:
[0178] SEQ ID NO.3: 5'-GGCGTTTTGAACAAACCGTC-3',...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com