Noninvasive prenatal bioinformatics detection system and method as well as application
A bioinformatics and detection method technology, applied in the field of non-invasive prenatal bioinformatics detection system, can solve the problem of inability to calculate the fetal concentration of female fetuses, and achieve the effect of reducing errors and complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1 Non-invasive prenatal bioinformatics detection device
[0078] This embodiment provides a non-invasive prenatal bioinformatics detection device, which specifically includes the following components:
[0079] (1) Original data acquisition unit: used to perform whole-genome sequencing on the extracted cfDNA from the peripheral blood of pregnant women to obtain the original data;
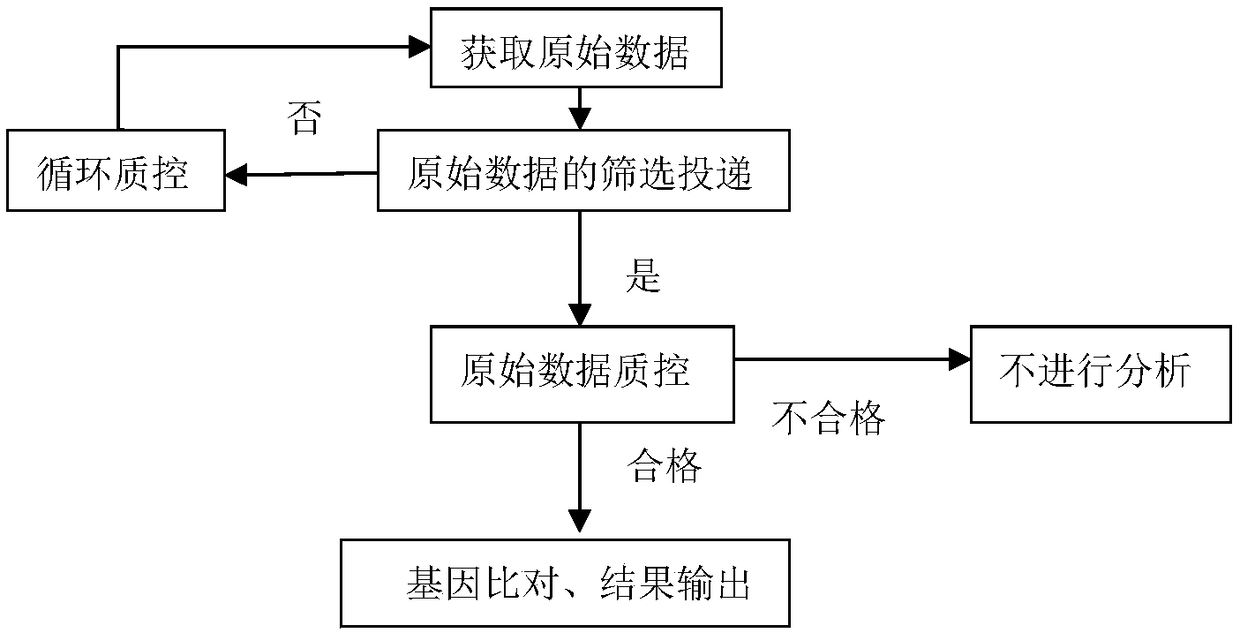
[0080] (2) Raw data screening and delivery unit: used for automatic monitoring, to determine whether the original data has completely completed the sequencing process and obtained the original data, and whether the sample configuration file is ready, if the result is "yes", enter the original data quality control unit; if If the result is "No", return to the original data acquisition unit, and enter the original data quality control unit until the result of the automatic monitoring program is "Yes";
[0081] (3) Raw data quality control unit: used to split the raw data with the result of "Yes"...
Embodiment 2
[0086] This embodiment provides a non-invasive prenatal bioinformatics detection method, using the device in embodiment 1, the process is as follows Figure 3-4 As shown, including the following steps:
[0087] (1) Obtain the original data: extract the cfDNA sample of the pregnant woman's peripheral blood, and then use the existing nextseq500 sequencer to build the database and sequence the sample.
[0088] (2) Screening and delivery of the original data: Before the sequencing on the computer, the automatic monitoring program must be started in advance to monitor whether the original data is completely off the computer and whether the samplesheet file used for data classification already exists, whether the delivery task can be started, etc.
[0089] The flow of the automatic monitoring program is as follows: First, enter the Linux command line mode, run the Linux screen command to create an analysis window, and directly run the monitoring shell script in the newly created window to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com