Third-generation data correction method based on DNA variation detection
A technology of mutation detection and data correction, applied in the field of bioinformatics, can solve problems such as high cost and high error rate, and achieve the effect of reducing cost, improving accuracy and facilitating data analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
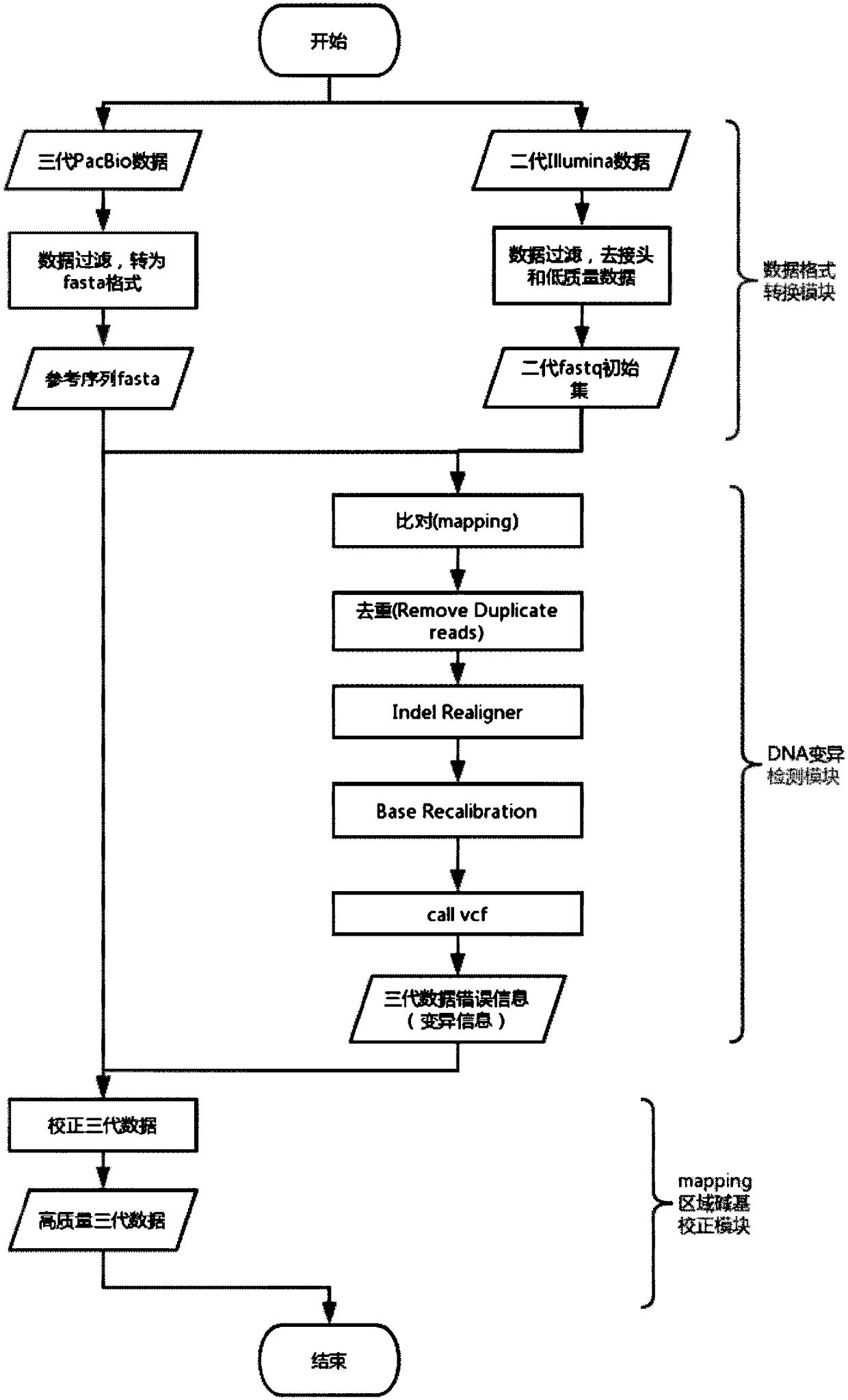
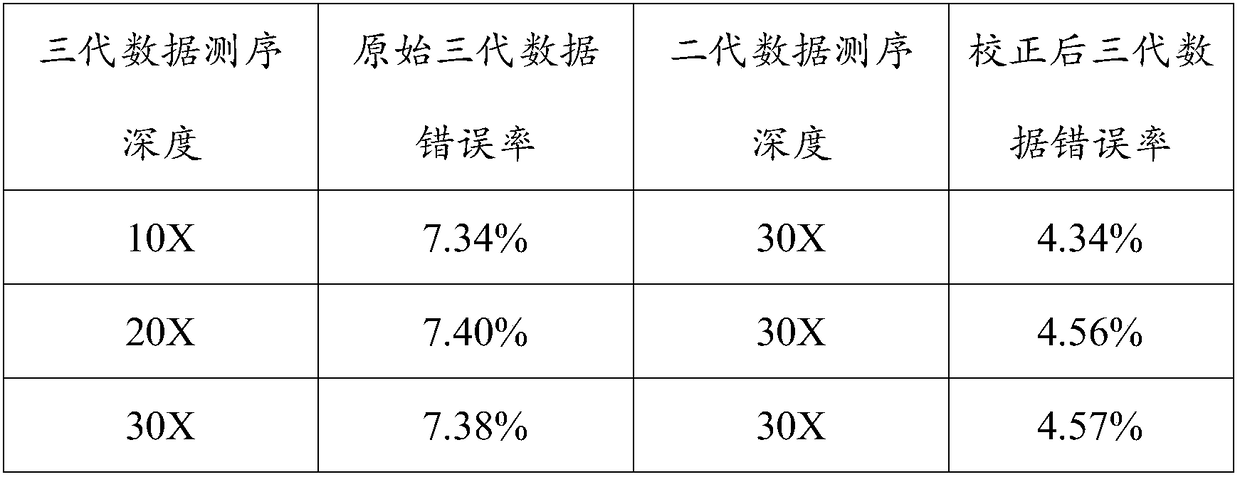
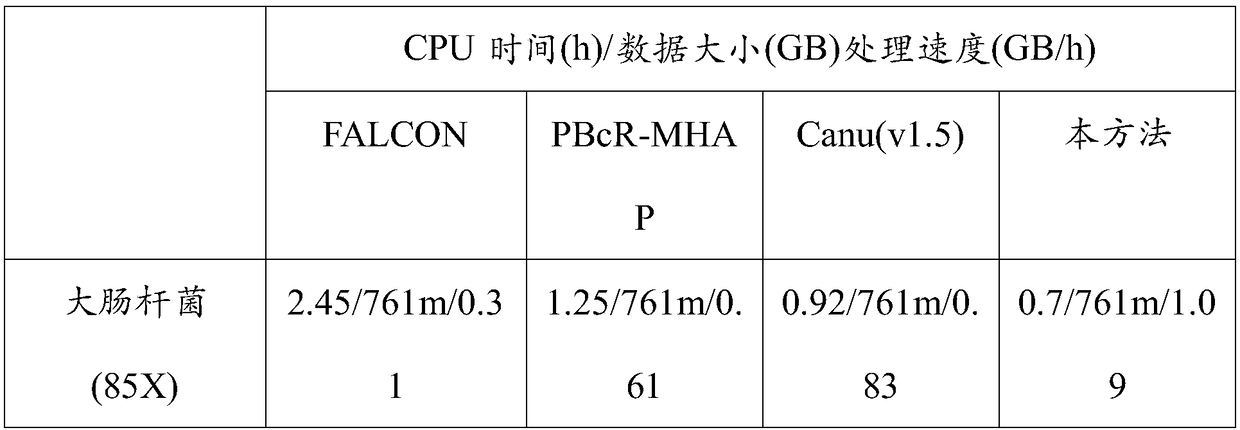
[0029] The third-generation data used in the test is the 85X Escherichia coli (Escherichia coliK12MG1655Methylome) sequencing data provided by PacBio (download address: https: / / github.com / PacificBiosciences / DevNet / wiki / Datasets), the second-generation data used is from NCBI’s sra The 290X Escherichia coli Illumina (Escherichia coli K12MG1655Methylome) sequencing data downloaded from the database with the number ERR022075, the selected reference genome is the standard reference gene of Escherichia coli K12MG1655 downloaded from the Genome database of NCBI, (download address: https: / / www .ncbi.nlm.nih.gov / genome / 167?genome_assembly_id=161521).
[0030] Set the coverage gradient of the PacBio data to 10X, 20X, and 30X respectively, and set the coverage gradient of the Illumina data to 30X. First compare the PacBio data to the reference genome, count the number of bases inserted and deleted in the mapping information in column 6 of the sam file, and calculate the sequencing error ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com