A kind of hemi-m methylation modification primer and application thereof
A methylation and hydroxymethylation technology, applied in the field of molecular biology, has achieved huge market value, solved the effect of comparative analysis of library DNA itself, and accurate methylation site analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] (1) A methylation modification primer, the primer sequence is shown in SEQ ID NO.1:
[0054] Said SEQ ID NO.1 is as follows:
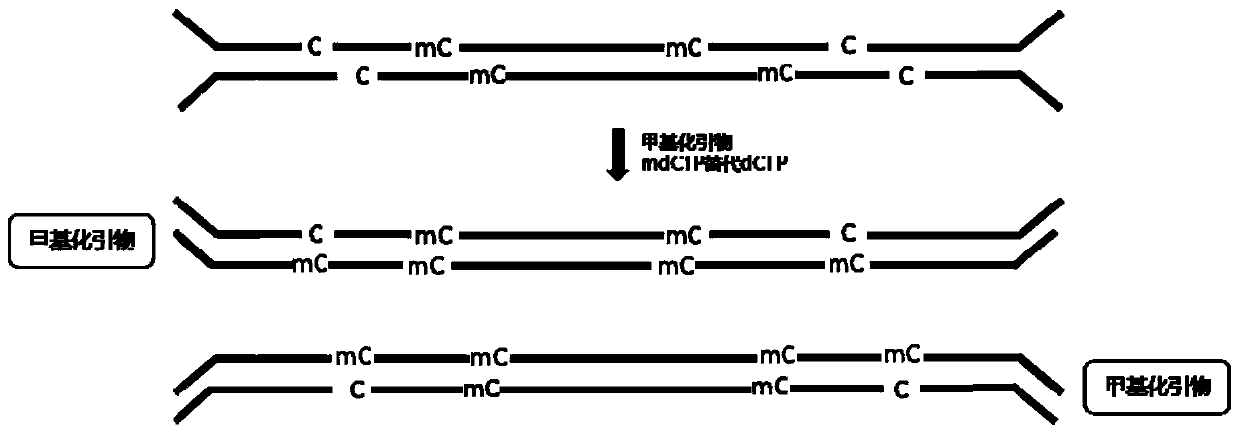
[0055] Primer 1: 5'GA / mdC / TGGAGTT / mdC / AGA / mdC / GTGTG 3'
[0056] The primer modification method is as follows:
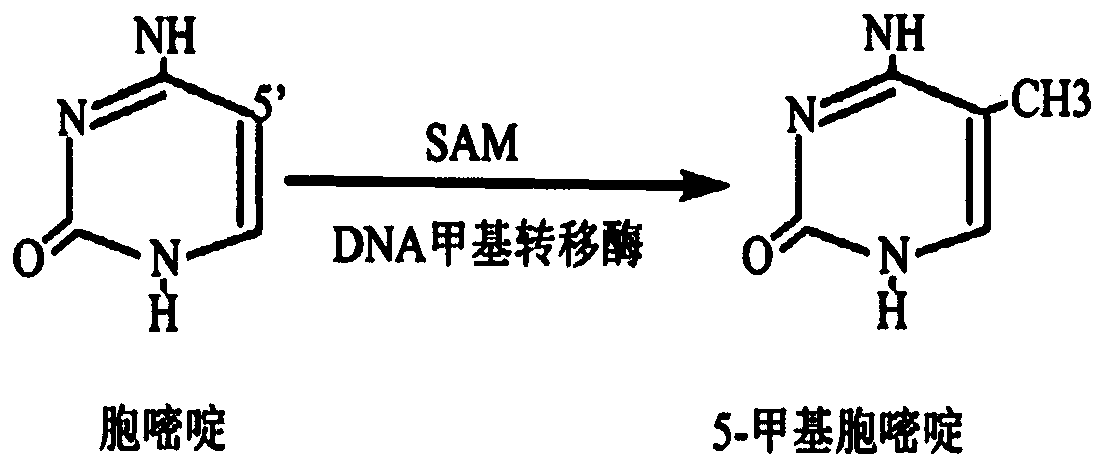
[0057] In the above methylated primer sequences, all cytosine (C) was modified to 5-methylcytosine (mdC) during synthesis.
[0058] (2) Sample processing:
[0059] If the sample to be processed is gDNA, use the fragmentation enzyme of NEB Company to fragment 2 μg of sample gDNA, and select 100-250 bp DNA fragments by magnetic beads or gel cutting method. Fragmentase treatment conditions: 37°C, 43min, The DNA after fragment selection was dissolved in 30 μL deionized water;
[0060] If the processed sample is cfDNA, it can be directly used in the subsequent experimental process.
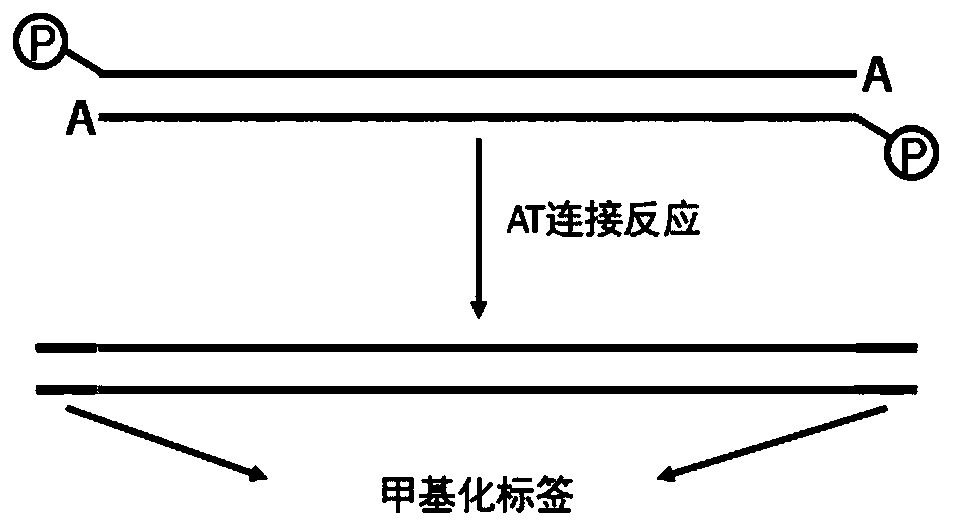
[0061] (3) Use the kit NEB library preparation kit for DNA end repair, add A to the 3' end, and ligate with methylated adapters.
[0062] (4) Cytosine (C...
Embodiment 2
[0079] (1) A methylation tag, the nucleotide sequences of the positive and negative strands of the methylation tag are shown in SEQ ID NO.2 and SEQ ID NO.3:
[0080] SEQ ID NO.2 is Barcode F: 5'NNNNNNNNTGAAT 3'
[0081] SEQ ID NO.3 is Barcode R: 5' -TT / mdC / ANNNNNNNNN 3’
[0082] Among them, Barcode F is reverse complementary to Barcode R, and the phosphate modification at the 5' end of Barcode R When the two strands are annealed, Barcode F hangs thymine at the 3' end and does not undergo phosphorylation at the 5' end.
[0083] Anneal Barcode F and Barcode R to label Barcode 1. Barcode 1 is a double-stranded Oligo. The sequence of each strand is the same as SEQ ID NO.2 and NO.3, except that the two strands are complementary to form a double-stranded structure.
[0084] (2) A methylation modification primer, the primer sequence is as follows:
[0085] SEQ ID NO.1 is Primer 1: 5'GA / mdC / TGGAGTT / mdC / AGA / mdC / GTGTG 3'
[0086] The primer modification method is as follows:
[...
Embodiment 3
[0113] (1) A tag, the nucleotide sequence of which is shown in SEQ ID NO.4:
[0114] SEQ ID NO.4 is Oligo: 5' -CATAG / dU / NNNNNNNNCTATGT 3’
[0115] The 5' end of Oligo is phosphorylated, and the five bases at the 5' end are reverse complementary to the 3' end. After annealing, a stem-loop structure is formed, and the 3' end overhangs the thymine label Barcode 2, as shown in Barcode 2 The nucleotide sequence sequence is SEQ ID NO.4, only self-complementary to form a stem-loop structure after annealing:
[0116] (2) A methylation modification primer, the primer sequence is as follows:
[0117] SEQ ID NO.1 is Primer 1: 5'GA / mdC / TGGAGTT / mdC / AGA / mdC / GTGTG 3'
[0118] The primer modification method is as follows:
[0119] In the above methylated primer sequences, all cytosine (C) was modified to 5-methylcytosine (mdC) during synthesis.
[0120] (3) Sample processing:
[0121] If the sample to be processed is gDNA, use the fragmentation enzyme of NEB Company to fragment 2 μg of s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com