Genome methylation library, preparation method and application thereof
A methylation and genome technology, applied in the field of genome methylation library and its preparation, can solve the problems of base imbalance, reduce the comparison rate of reads containing non-methylated cytosine, and cannot completely cover the CpG region, etc. Achieve the effect of improving the quality of sequencing and improving the base balance of the library
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0034] The method for preparing a genome methylation library according to an embodiment of the present invention includes the following steps S1-S5:
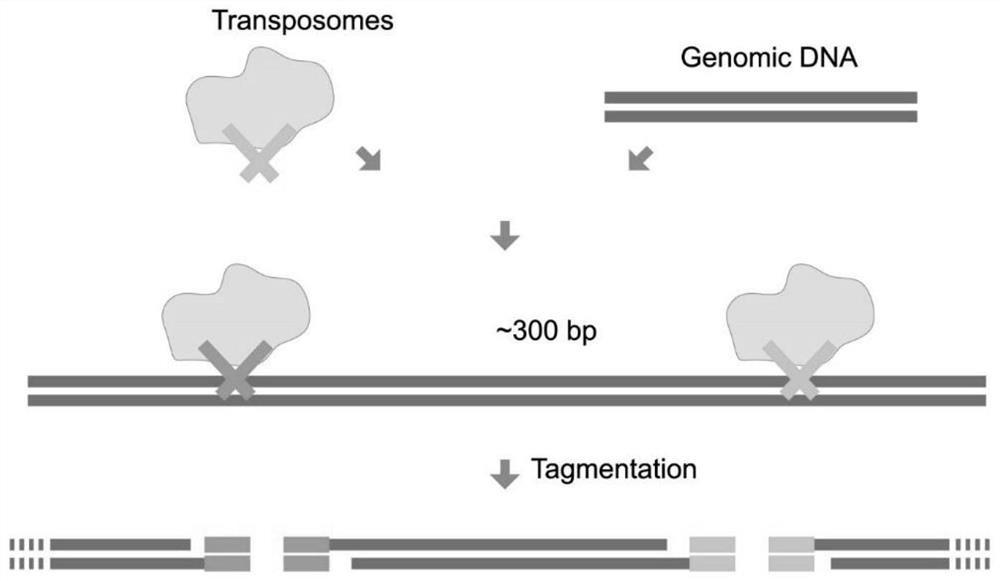
[0035] S1, providing a transposase complex (transposome), the transposase complex is composed of a transposable adapter and a transposase, and the transposable adapter contains a transposon sequence;
[0036] S2. Using a transposase complex to fragment genomic DNA (tagmentation) to obtain a fragmented product with a transposable adapter and a gap;
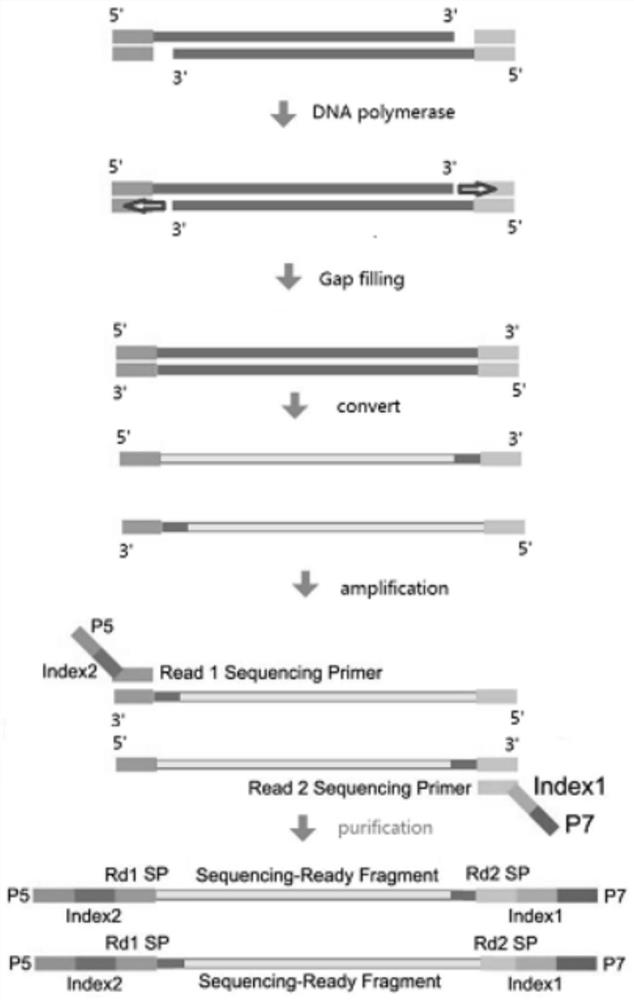
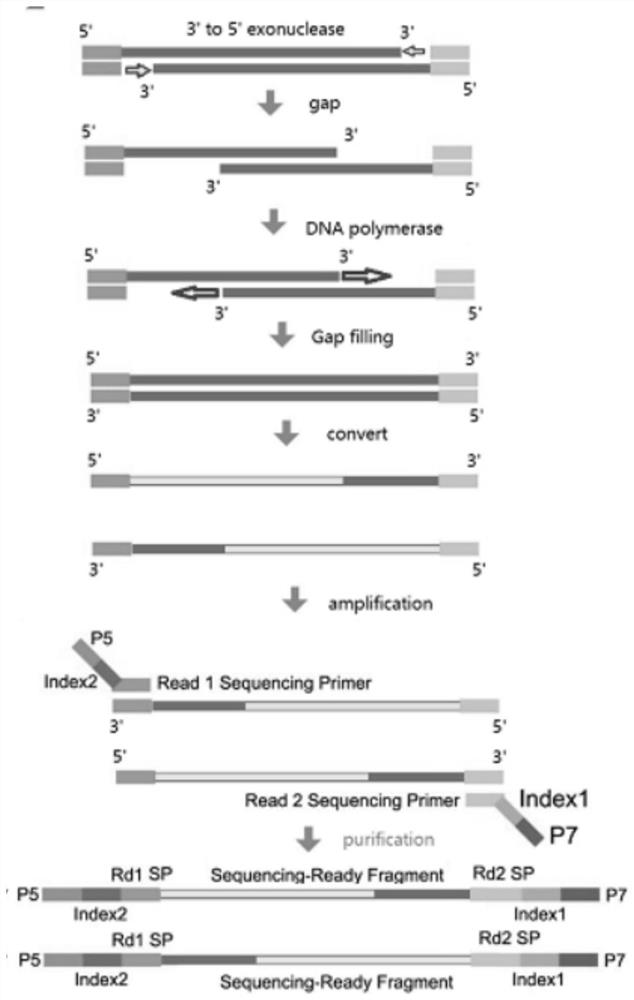
[0037] S3, using dNTPs that replace dCTP with mdCTP to perform gap filling (gap filling) on the fragmented product;
[0038] S4, converting the fragmented product after the gap filling, so that the unmethylated cytosine is converted into uracil;
[0039] S5. Perform PCR amplification (amplification) on the transformed fragmented product to obtain a genome methylation library.
[0040] The present invention fragments the genomic DNA through the transposase complex and adds adapter...
Embodiment 1
[0058] Example 1 Genome methylation library construction mediated by Tn5 transposase
[0059] (1) Sample preparation: HEK293 cells were taken, and DNA was extracted using a cell genome DNA extraction kit. The resulting nucleic acids were dissolved in 50 μL TE buffer, and the concentration was measured using a Qubit 3.0 fluorometer. Finally, 50 ng DNA was taken for library construction according to the following operation steps.
[0060] (2) Tn5 processing mark:
[0061] a. Synthesize a nucleic acid fragment containing a 19bp transposase recognition sequence (the detailed sequence is shown in the following SEQ ID NO: 1, SEQ ID NO: 2 and SEQ ID NO: 3), wherein part of the cytosine in sequence B and sequence C is formazan Modified cytosine ( C ). In sequence B and sequence C, "N" represents any base A, T, C or G, wherein cytosine is methylated modified cytosine.
[0062] Linker sequence A: CTGTCTCTTATACACATCT (SEQ ID NO: 1);
[0063] Linker sequence B: T C GT C GG C AG ...
Embodiment 2
[0084] Example 2 Construction of genome methylation library mediated by Tn5 transposase and nick-digested
[0085] (1) Sample preparation: HEK293 cells were taken, and DNA was extracted using a cell genome DNA extraction kit. The resulting nucleic acids were dissolved in 50 μL TE buffer, and the concentration was measured using a Qubit 3.0 fluorometer. Finally, 50 ng DNA was taken for library construction according to the following operation steps.
[0086] (2) Tn5 processing mark:
[0087]a. Synthesize a nucleic acid fragment containing a 19bp transposase recognition sequence (the detailed sequence is shown in the following SEQ ID NO: 1, SEQ ID NO: 2 and SEQ ID NO: 3), wherein part of the cytosine in sequence B and sequence C is formazan Modified cytosine ( C ). In sequence B and sequence C, "N" represents any base A, T, C or G, wherein cytosine is methylated modified cytosine.
[0088] Linker sequence A: CTGTCTCTTATACACATCT (SEQ ID NO: 1);
[0089] Linker sequence B: T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com