Hemi-M methylation-modified primer and application thereof
A methylation and hydroxymethylation technology, applied in the field of molecular biology, to achieve broad application prospects, reduce complex processes, and accurately analyze methylation sites.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] (1) A methylation modification primer, the primer sequence is shown in SEQ ID NO.1:
[0054] Said SEQ ID NO.1 is as follows:
[0055] Primer 1: 5'GA / mdC / TGGAGTT / mdC / AGA / mdC / GTGTG 3'
[0056] The primer modification method is as follows:
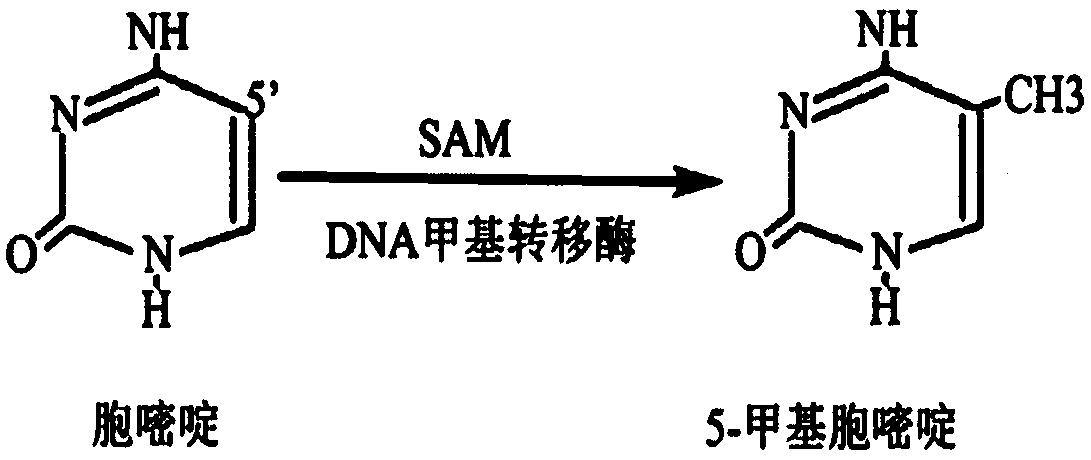
[0057] In the above methylated primer sequences, all cytosine (C) was modified to 5-methylcytosine (mdC) during synthesis.
[0058] (2) Sample processing:
[0059] If the sample to be processed is gDNA, use the fragmentation enzyme of NEB Company to fragment 2 μg of sample gDNA, and select 100-250 bp DNA fragments by magnetic beads or gel cutting method. Fragmentase treatment conditions: 37°C, 43min, The DNA after fragment selection was dissolved in 30 μL deionized water;
[0060] If the processed sample is cfDNA, it can be directly used in the subsequent experimental process.
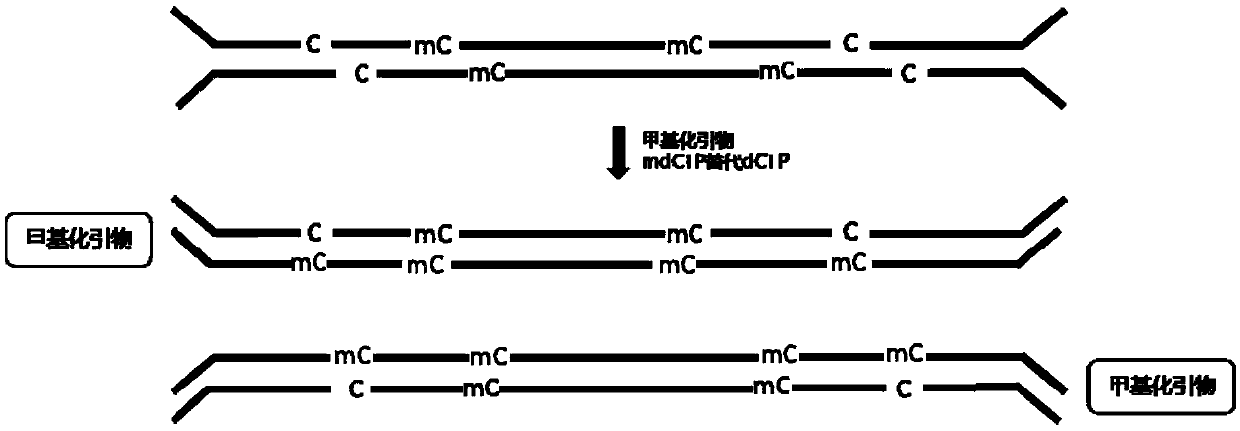
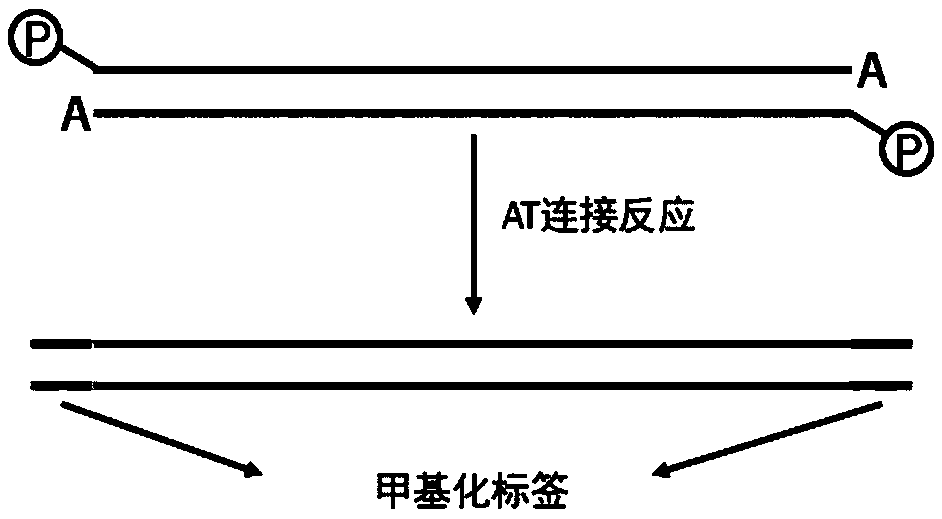
[0061] (3) Use the kit NEB library preparation kit for DNA end repair, add A to the 3' end, and ligate with methylated adapters.
[0062] (4) Cytosine (C...
Embodiment 2
[0079] (1) A methylation tag, the nucleotide sequences of the positive and negative strands of the methylation tag are shown in SEQ ID NO.2 and SEQ ID NO.3:
[0080] SEQ ID NO.2 is Barcode F: 5'NNNNNNNNTGAAT 3'
[0081] SEQ ID NO.3 is Barcode R: 5' -TT / mdC / ANNNNNNNNN 3’
[0082] Among them, Barcode F is reverse complementary to Barcode R, and the phosphate modification at the 5' end of Barcode R When the two strands are annealed, Barcode F hangs thymine at the 3' end and does not undergo phosphorylation at the 5' end.
[0083] Anneal Barcode F and Barcode R to label Barcode 1. Barcode 1 is a double-stranded Oligo. The sequence of each strand is the same as SEQ ID NO.2 and NO.3, except that the two strands are complementary to form a double-stranded structure.
[0084] (2) A methylation modification primer, the primer sequence is as follows:
[0085] SEQ ID NO.1 is Primer 1: 5'GA / mdC / TGGAGTT / mdC / AGA / mdC / GTGTG 3'
[0086] The primer modification method is as follows:
[...
Embodiment 3
[0113] (1) A tag, the nucleotide sequence of which is shown in SEQ ID NO.4:
[0114] SEQ ID NO.4 is Oligo: 5' -CATAG / dU / NNNNNNNNCTATGT 3’
[0115] The 5' end of Oligo is phosphorylated, and the five bases at the 5' end are reverse complementary to the 3' end. After annealing, a stem-loop structure is formed, and the 3' end overhangs the thymine label Barcode 2, as shown in Barcode 2 The nucleotide sequence sequence is SEQ ID NO.4, only self-complementary to form a stem-loop structure after annealing:
[0116] (2) A methylation modification primer, the primer sequence is as follows:
[0117] SEQ ID NO.1 is Primer 1: 5'GA / mdC / TGGAGTT / mdC / AGA / mdC / GTGTG 3'
[0118] The primer modification method is as follows:
[0119] In the above methylated primer sequences, all cytosine (C) was modified to 5-methylcytosine (mdC) during synthesis.
[0120] (3) Sample processing:
[0121] If the sample to be processed is gDNA, use the fragmentation enzyme of NEB Company to fragment 2 μg of s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com