Primer for detecting pathogenic microorganism and multiple PCR using the same
A pathogenic microorganism and multiple amplification technology, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, recombinant DNA technology, etc., can solve the cumbersome and time-consuming problems, the inability to quickly and accurately diagnose, and the lack of specific high-quality diagnostic serum and other problems, to achieve broad application prospects and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Embodiment 1, intra-species and inter-species mixed primer PCR
[0052] 1. Template Preparation
[0053] Pick a single colony from a petri dish and inoculate it in a liquid medium for culture, take an appropriate amount of bacterial solution and put it in a boiling water bath for 10 minutes, centrifuge at 13000rpm for 10 minutes, and take the supernatant as a template.
[0054] 2. Selection of virulence genes and specific primers of enteric pathogens
[0055] The following genes were selected by consulting the literature and the GenBank database (see Table 2): rfbE, eaeA of EHEC, lt, st of ETEC, eaeA, bfpA of EPEC, ipaH of EIEC, o1ag, ct of Vibrio cholerae O1 serogroup Eltor biotype , rtx, o1ag, ct of O1classic, o139ag, rtx of O139 group, tl, tdh, trh of Vibrio parahaemolyticus, mapA, bp, cdt of Campylobacter jejuni, iroB, rfbS of Salmonella.
[0056] Primers were designed according to the conserved region of the gene, the length of the product was 300-500 bp, and the...
Embodiment 2
[0077] Embodiment 2, multiplex PCR incorporation of fluorescein control without the output of fluorescein
[0078] The difference in the yield of the amplified product was compared with and without the addition of fluorescein in the reaction system.
[0079]Reaction system: 25μl, 2.5μl of 10×PCR buffer, 0.5μl of three dNTPs (10mM, without dTTP), 4μl of dTTP (1mM), 0.5μl of cy5-dUTP (1mM), 1u of taq enzyme (2u / μl), Primer (1.6 μM) 2.5 μl, template 0.5 μl, deionized water 14 μl. The reaction conditions are the same as single weight.
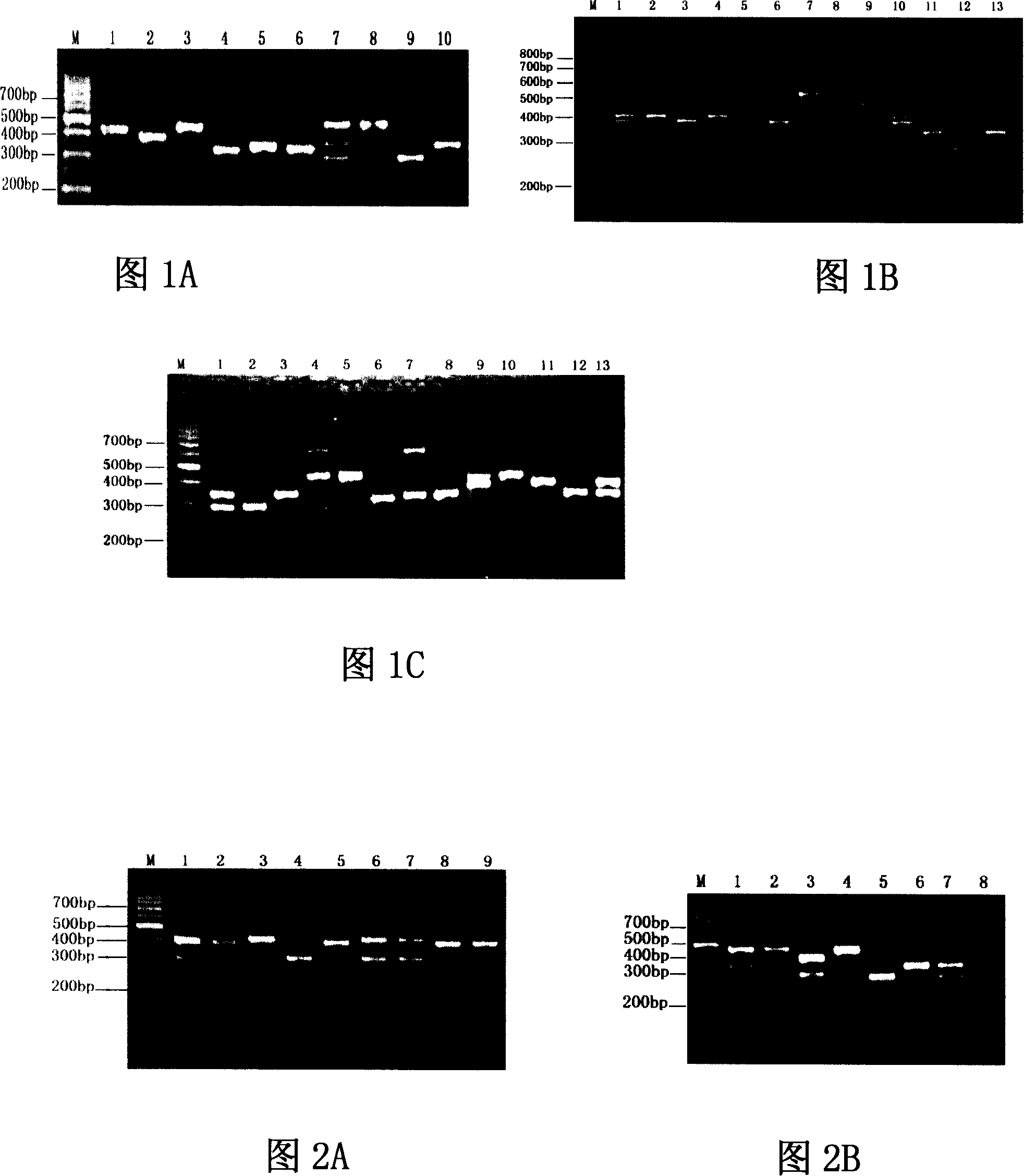
[0080] Results: The incorporation of fluorescein in the reaction system significantly reduced the amplification yield (see Figure 2A and Figure 2B).
Embodiment 3
[0081] Embodiment 3, the making and hybridization of gene chip
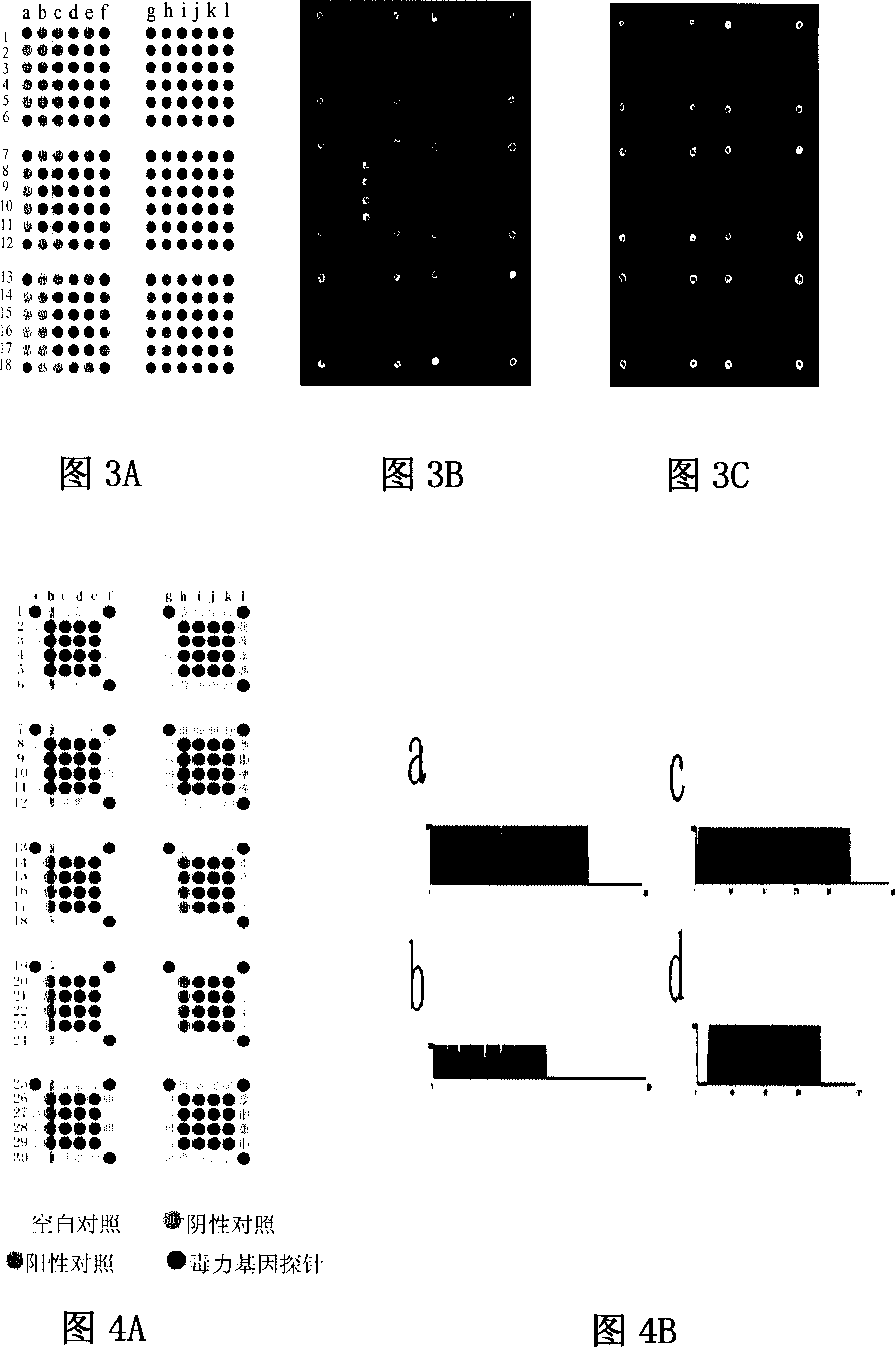
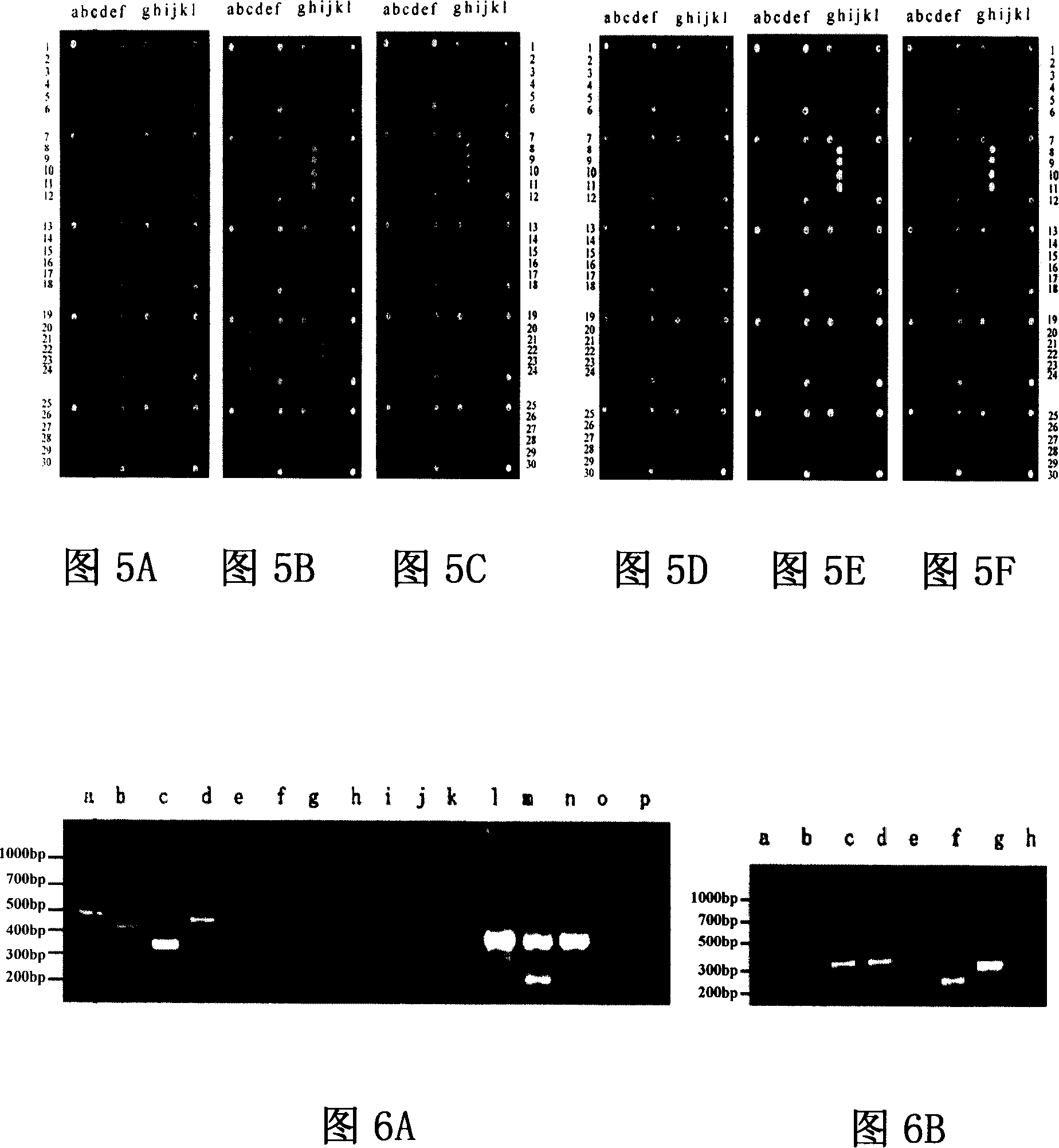
[0082] Use spotting instrument (GeneTAC Microarraying, Perkin Elmer Company, USA) to spot specific virulence gene fragments in a certain order on amino-modified glass slides, including six 6×6 matrices, each matrix consists of four positioning coordinate points and four probes, each probe longitudinally replicated four points. The chips were fixed by rehydration, UV cross-linking and high-temperature dry-baking. The lattice composition is shown in Figure 3A. Put the processed chip into a GenTAC Hybridization hybridization instrument, add 100 μl of pre-hybridization solution, agitate at 42° C. for 40 minutes, and wash with 0.1% SDS. Take 20 μl of the purified labeled sample, add 80 μl of hybridization solution, mix well, inject into the hybridization instrument, cover the chip probe sample array, hybridize at 42°C for 12 hours, wash unbound probes with cleaning solution, desalt, and dry in the air. The scanner ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com