Screening method for aptamers and aptamer specifically bond with pseudomonas aeruginosa
A Pseudomonas aeruginosa and nucleic acid aptamer technology, applied in biochemical equipment and methods, microbial measurement/inspection, instruments, etc., can solve the problem that the on-site detection technology of Pseudomonas aeruginosa has not attracted too much research attention , Pseudomonas aeruginosa cannot be detected and other problems, to achieve good detection ability, good specificity and sensitivity, and accurate detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
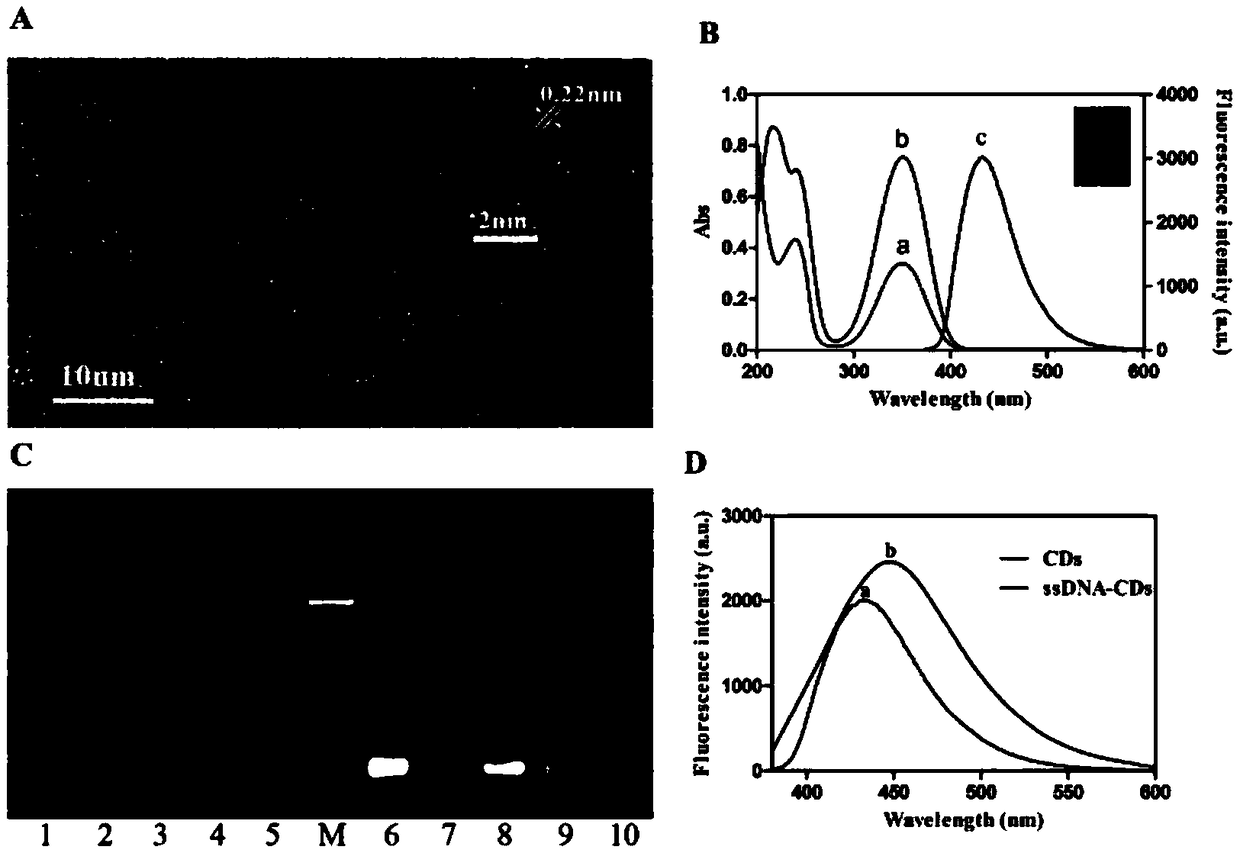
[0046] Example 1 Aptamer Screening
[0047] 1. Design random library sequences:
[0048] 5'-TGGACCTTGCGATTGCGATTGACAGC(40nt)GCAGACATGAGTCT-CAGGAC-3' has a random region of 40 nucleotides in the middle and a fixed region of 20 nucleotides at both ends. The total sequence length is 80 nucleotides.
[0049] The primer sequences used for screening are:
[0050] Upstream primer: TGGACCTTGCGATTGACAGC
[0051] Downstream primer 1: GTCCTGAGACTCATGTCTGC
[0052] Downstream primer 2: 5'-biotinGTCCTGAGACTCATGTCTGC
[0053] The above nucleic acid sequences were completed by Shanghai Sangon Biotechnology Co., Ltd.
[0054] 2. Using WB-SELEX technology to screen Pseudomonas aeruginosa aptamers
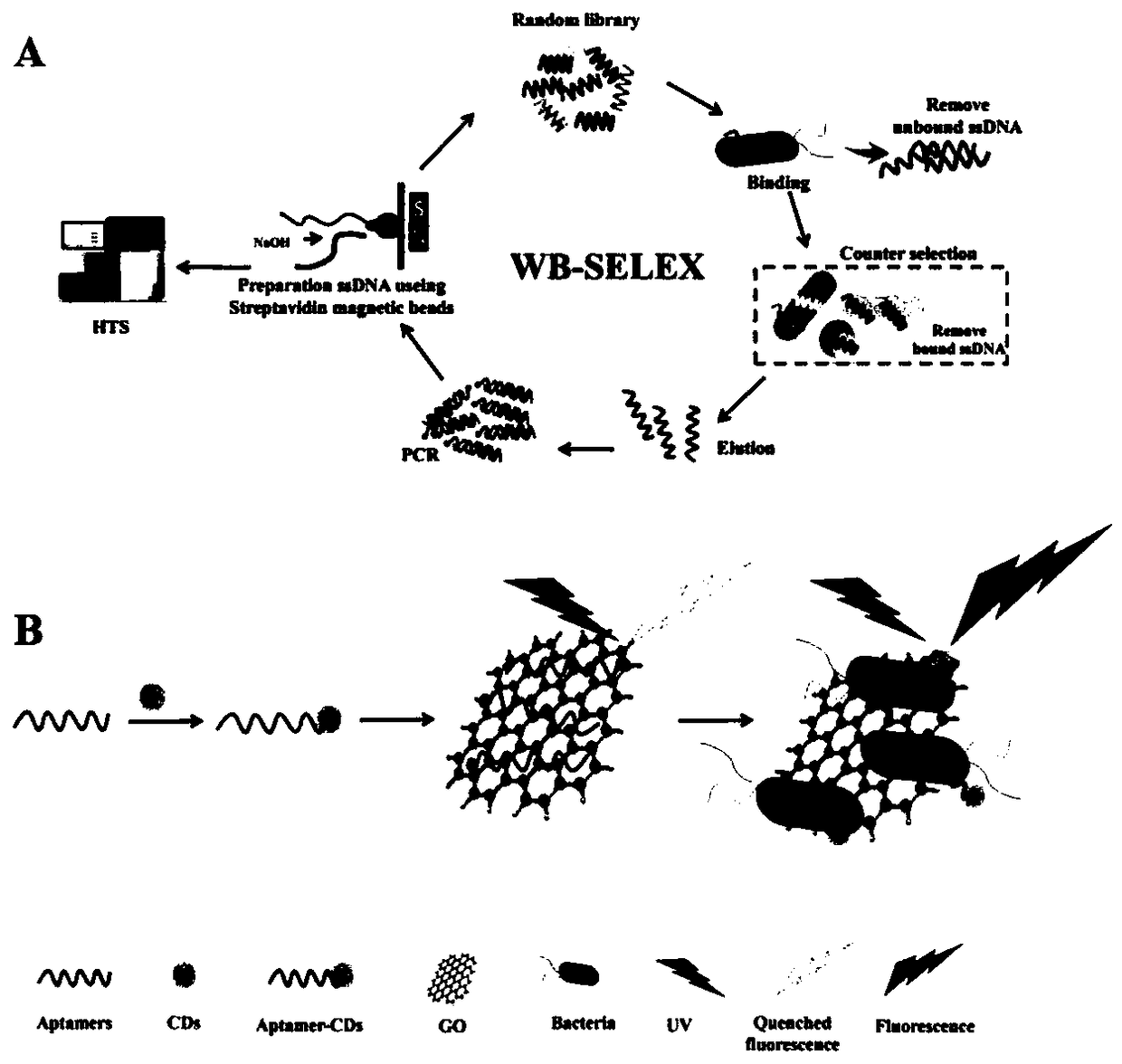
[0055] The screening process of Pseudomonas aeruginosa nucleic acid aptamer is as follows: figure 1 -A shown.
[0056] 1) The strains used for screening and their cultivation and collection:
[0057] Aptamer screening was performed using Pseudomonas aeruginosa ATCC 27853. E. coli ATCC25922,...
Embodiment 2
[0089] Embodiment 2 detects Pseudomonas aeruginosa
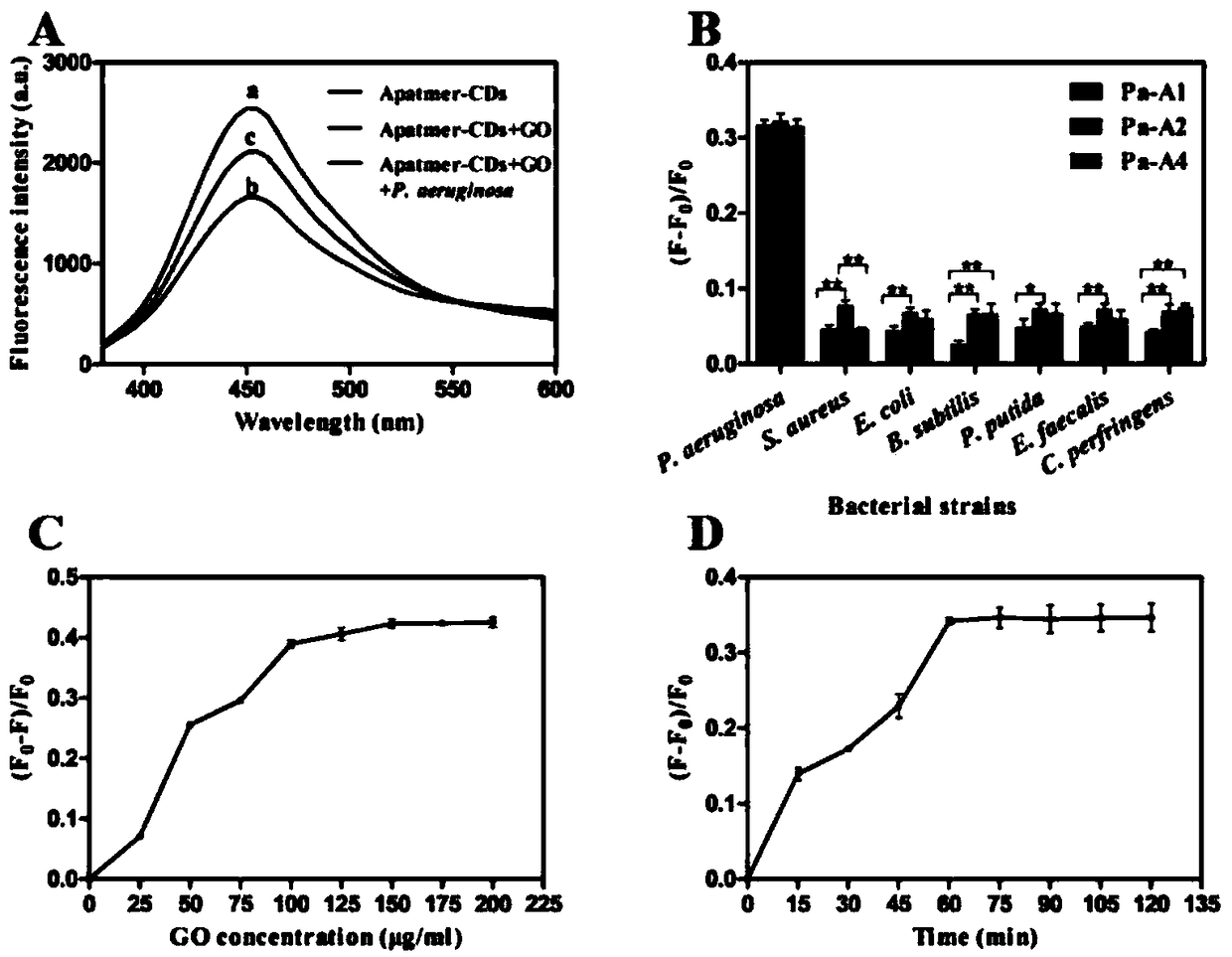
[0090] 1. Synthesize the aptamer-CD / GO biosensor first
[0091] CDs were synthesized using a hydrothermal method, and then 100 μL of CD solution (8 μg / mL) in 5 mM PBS buffer was mixed with 30 μl of N-(3-dimethylaminopropyl)-ethylcarbodiimide hydrochloride (EDC) (1 mg / μl) and N-hydroxysuccinimide (NHS) (1 mg / μl) to activate the carboxyl group, and sonicate for 30 min at 25°C. Afterwards, 10 μL of the sonicated mixture was added to 90 μL of candidate aptamers with modified amino groups at a concentration of 3.3 μM in 5 mM PBS buffer and incubated at 25 °C for 2 h to prepare aptamer-CD. 50 μL of GO (1 mg / mL) was added to 25 μL of the aforementioned aptamer-CDs solution (100 nM) to quench the fluorescence, and then the mixture was incubated at 37 °C for 1 hour to prepare the aptamer-CD / GO biosensor.
[0092] 2. Detection of Pseudomonas aeruginosa
[0093] 100 μL of Pseudomonas aeruginosa (1×10 7 CFU / ml) was added to the reac...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com