Method and device for calculating purity and chromosome ploidy of cancer sample
A chromosome ploidy, cancer technology, applied in the field of cancer research, can solve the problems of inconvenience, poor accuracy, lack of accuracy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
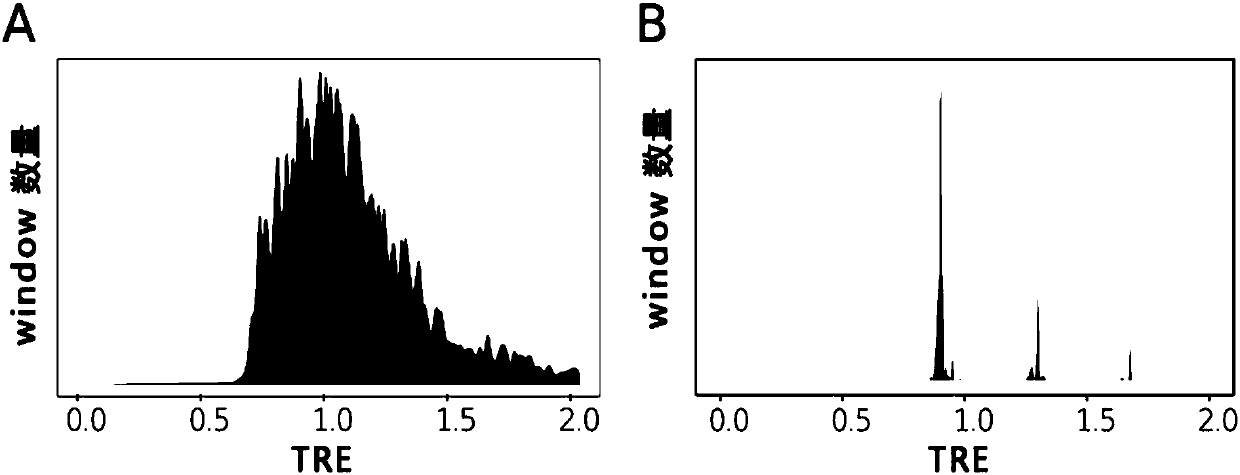
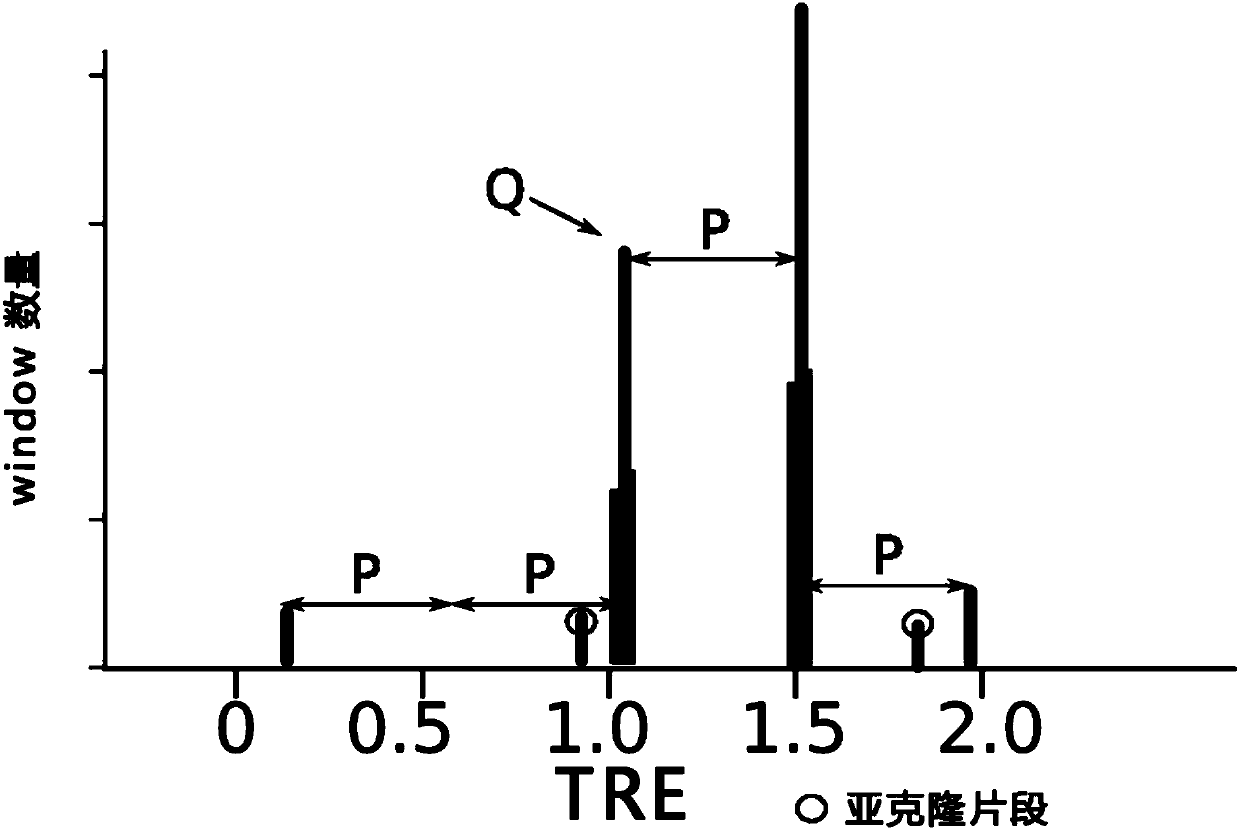
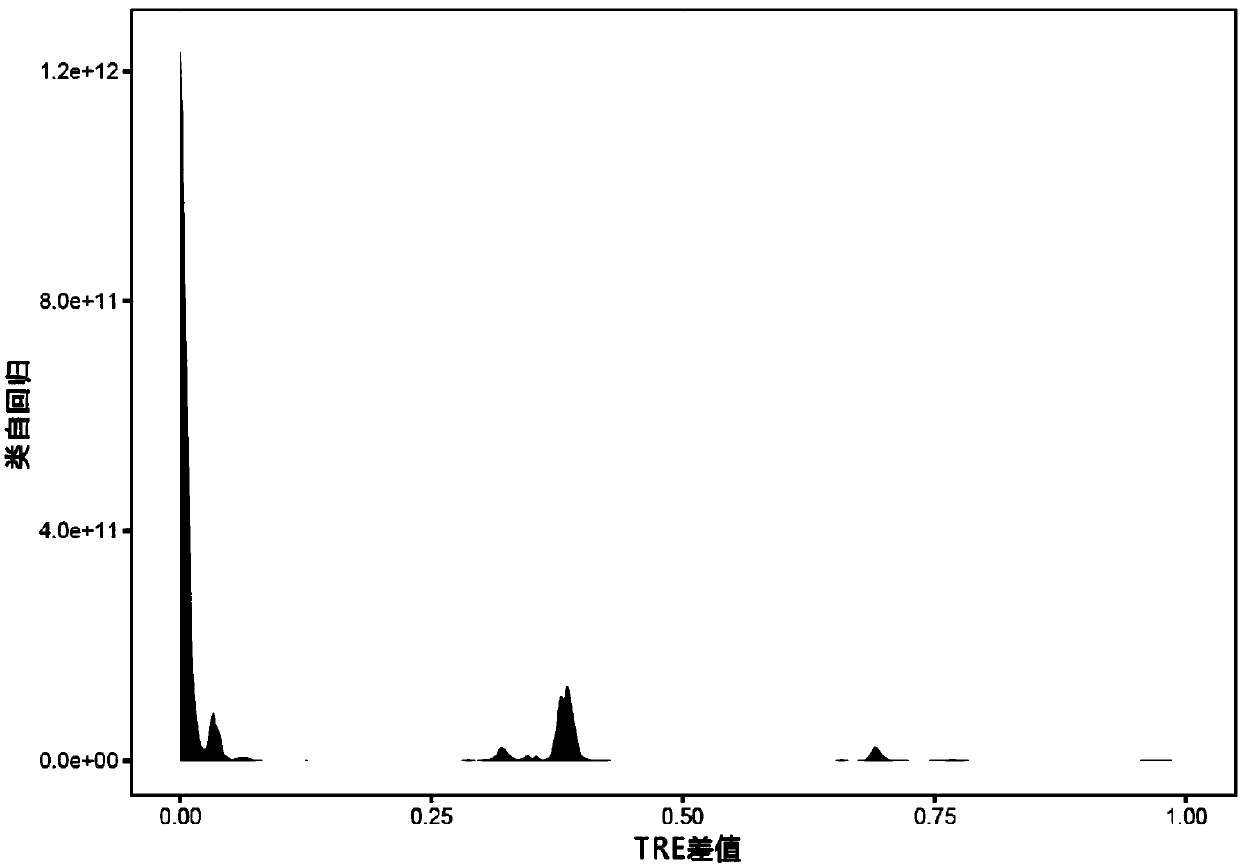
[0251] Example: According to the whole genome sequencing data of cancer tissue and normal tissue of sample TCGA-CM-4746, the purity and chromosomal ploidy of cancer samples were calculated using a hierarchical mixed Gaussian model.
[0252] 1. Collect sample data and download the whole genome sequencing data of TCGA-CM-4746-01A tumor samples and normal samples in TCGA. The bam file size of the cancer sample is 12.6G, and the bam file size of the normal sample is 10.1G. The bam file was processed into a fastq file with PICARD software. Align fastq to the reference genome hs37d5 using bwa mem to get bam files of new cancer samples and normal samples, the file sizes are 12.4G and 9.9G respectively.
[0253] 2. Download the vcf files of chromosomes 1 to 22 provided by the 1000genome project (ftp: / / ftp.1000genomes.ebi.ac.uk / vol1 / ftp / release / 20130502 / ), and use the SelectVariants method of GATK to extract the reference genome hs37d5 The BIALLELIC loci with an allele frequency grea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com