Tumor-specific gene identification method based on pls multi-perturbation integrated gene selection
A gene selection and identification method technology, applied in the interdisciplinary field of computing science and life science, can solve the problem of insufficient insight into the whole picture of complex genetic mechanisms, and achieve reliable molecular diagnosis and treatment, strong discrimination ability, and small length
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0046] The present invention will be further described below in combination with implementation modes and examples.
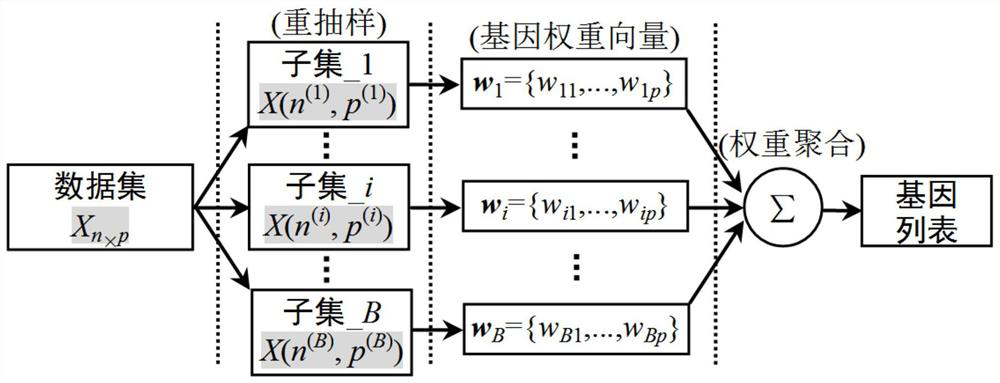
[0047] Such as figure 1 As shown, this embodiment provides a method for identifying tumor-specific genes based on PLS multi-perturbation integrated gene selection, including the following steps:
[0048] Step S1: Establish a multi-response variable PLS model, use the SIMPLS algorithm to solve the multi-response variable PLS model, and realize polygenic measurement based on PLS;
[0049] Step S2: Using the PLS-based multi-gene measurement method, under the framework of multi-perturbation integrated gene selection, perform gene selection based on PLS integration on the sample data, and obtain the gene list of the sample data;
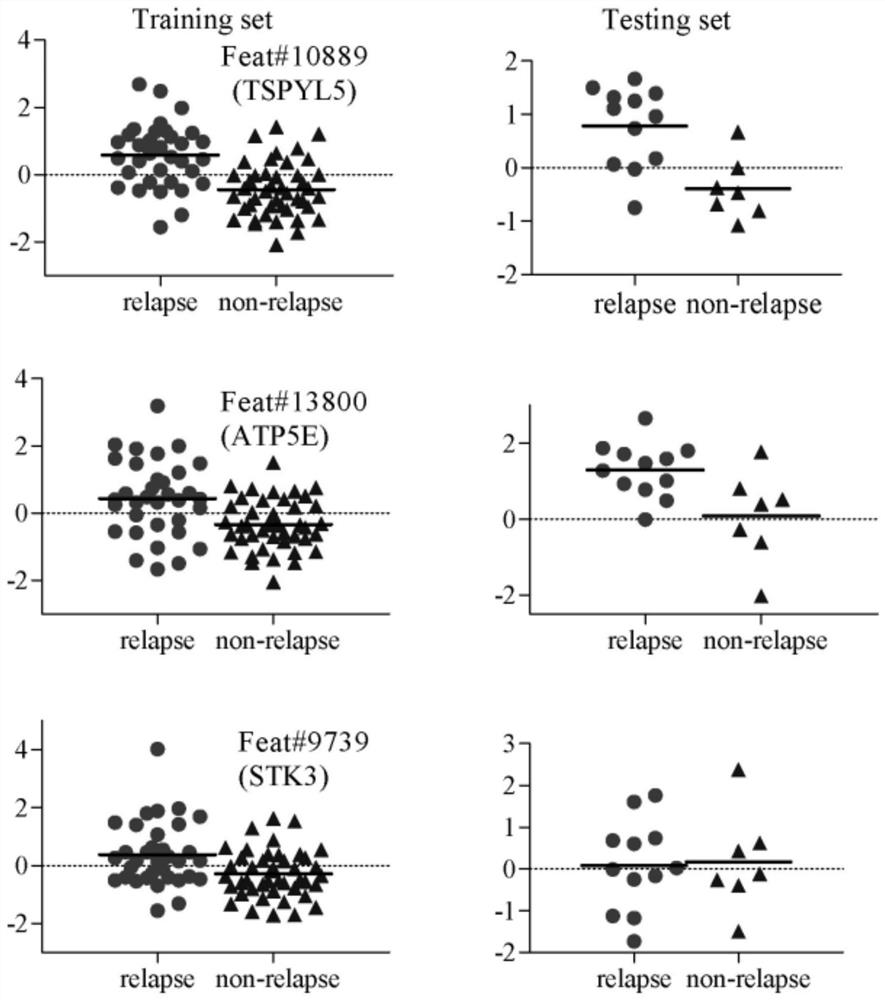
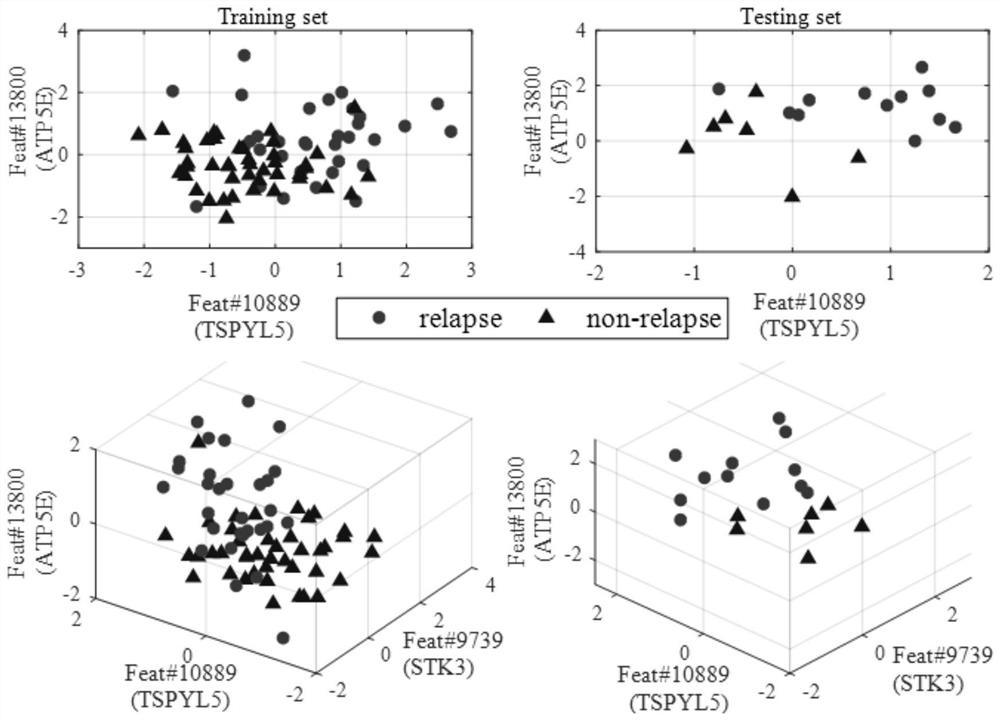
[0050] Step S3: Using the base classifier, identify the top k genes with the highest recognition rate from the above sorted gene list to form a tumor-specific gene subset.
[0051] In this example, gene selection is from the original hig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com