Optimization method of PIK3CA gene H1047R mutation digital PCR detection system and detection product
An optimization method and digital technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of low cfDNA content, insensitive and accurate detection of plasma cell-free DNA mutation information, etc., and achieve accurate results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
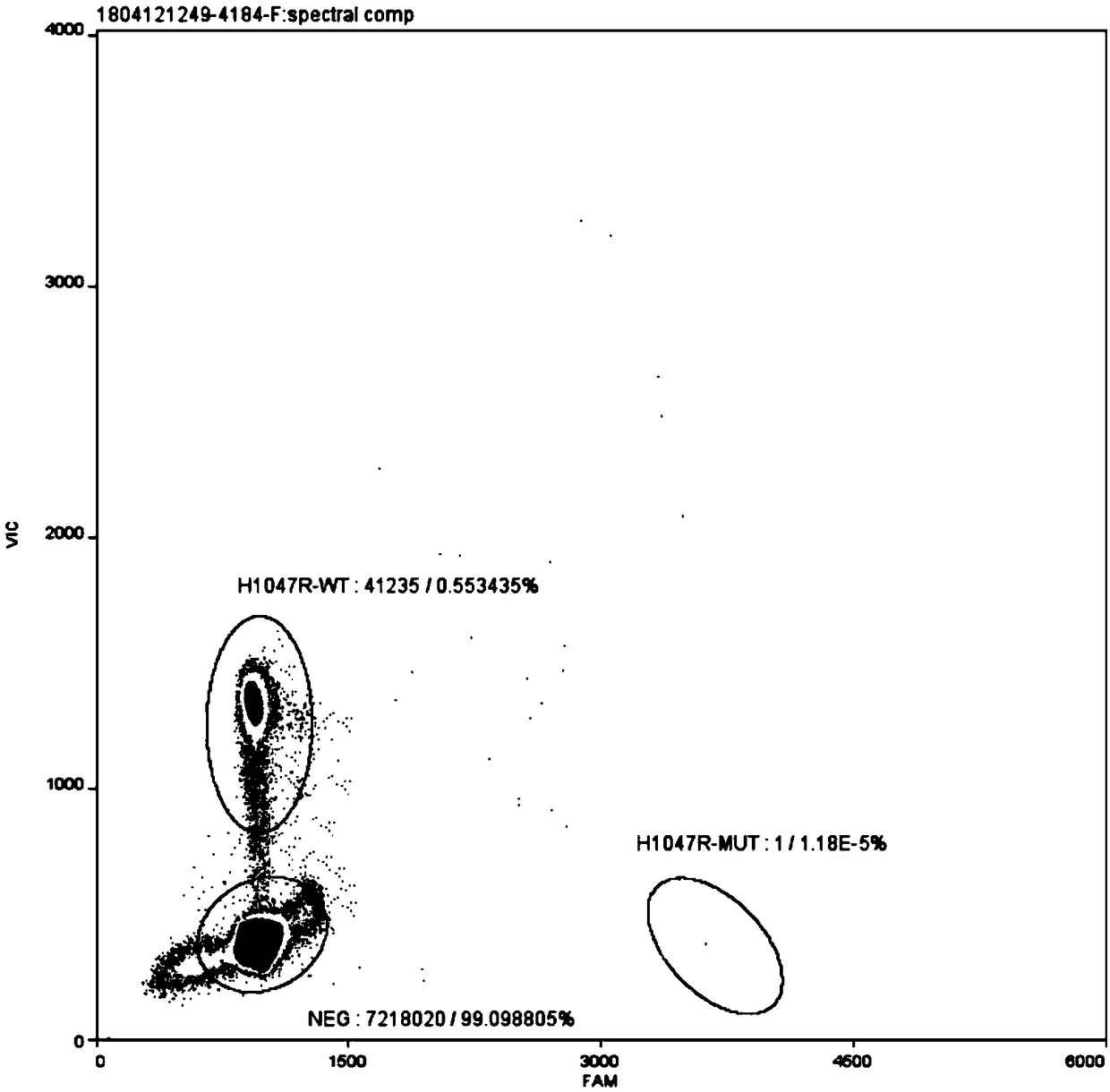
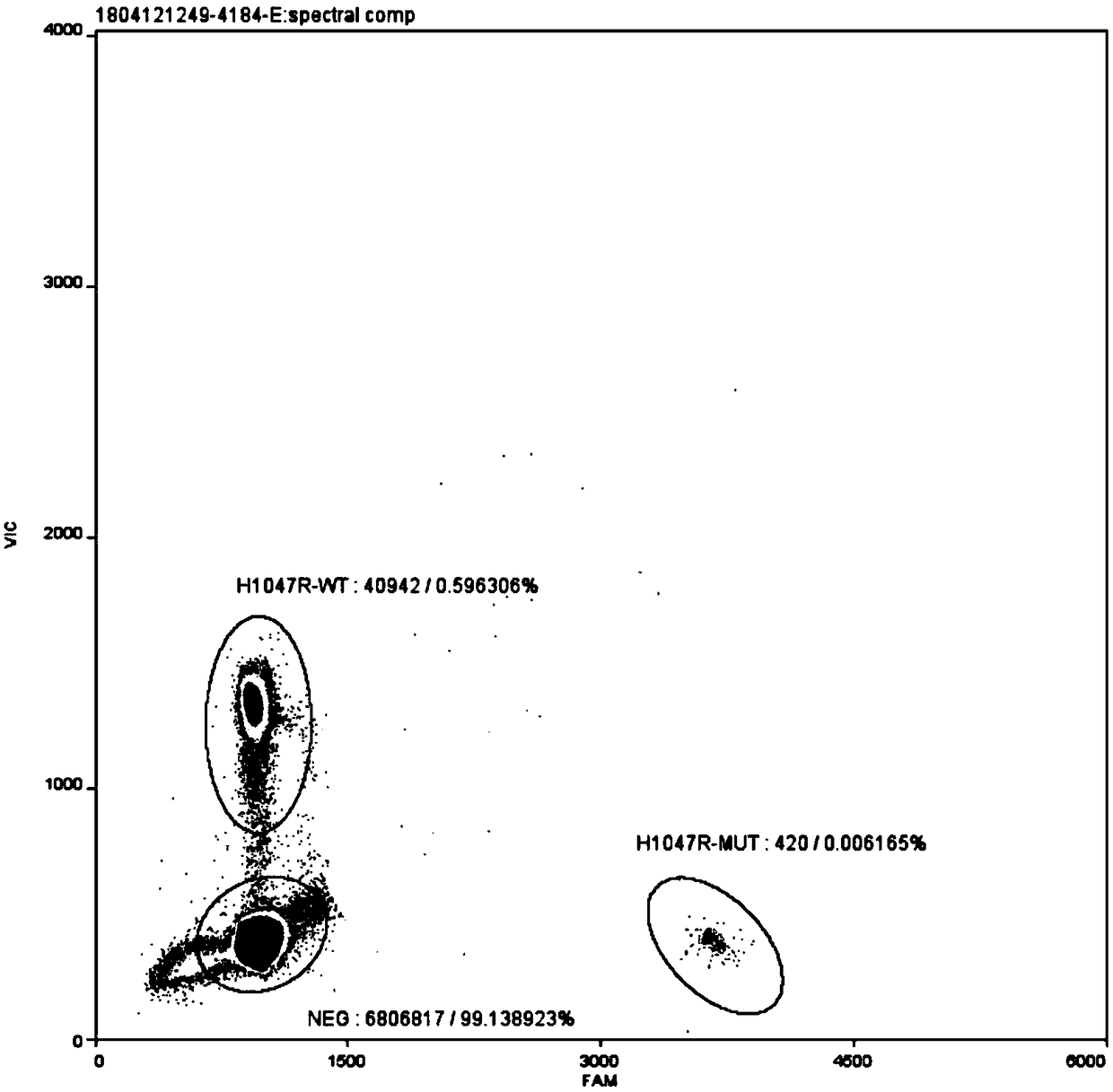
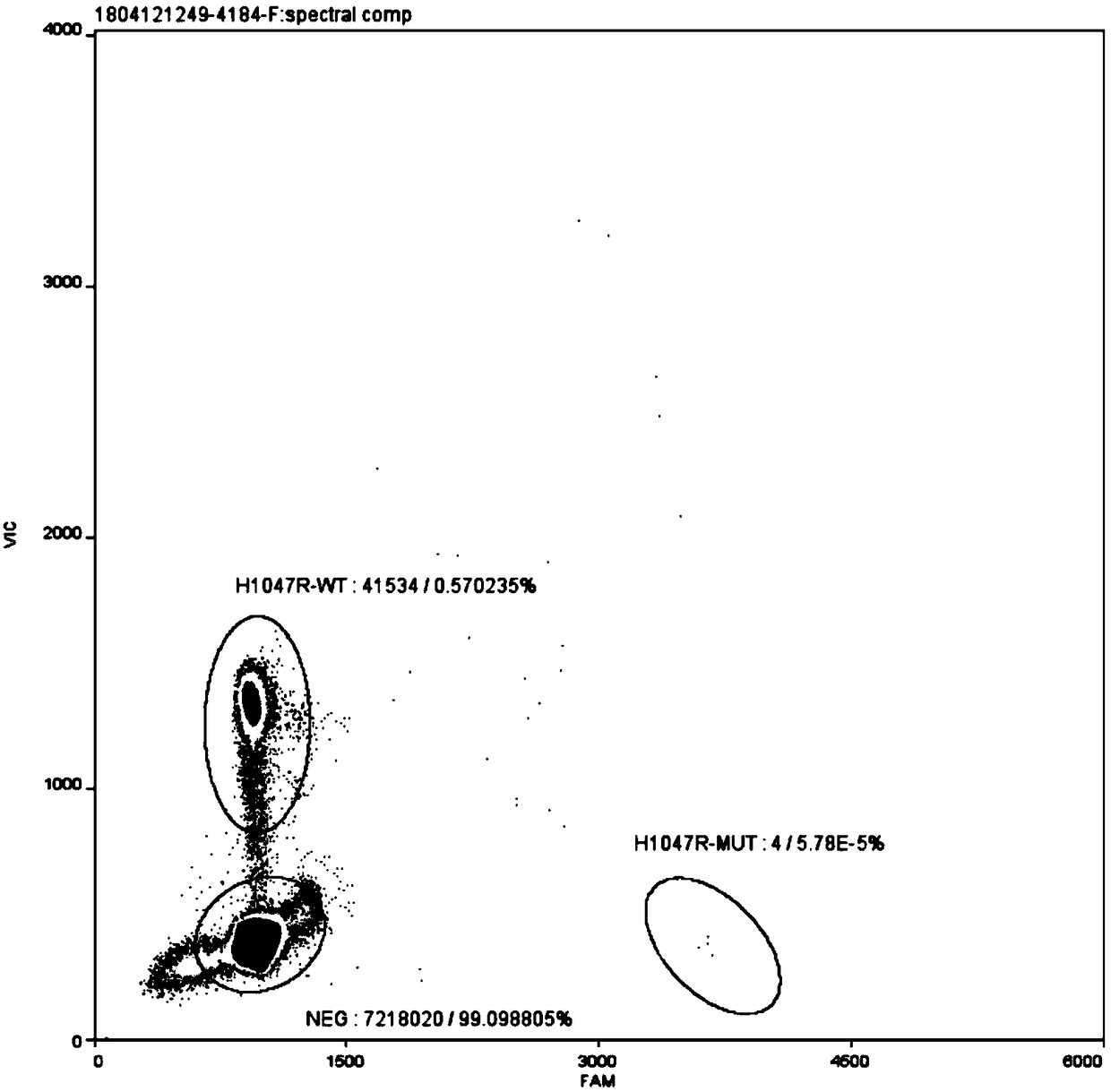
[0034] The PIK3CA gene H1047R mutation digital PCR detection kit of this embodiment includes upstream primer (PIK3CA-H1047R-F), downstream primer (PIK3CA-H1047R-R), wild-type probe (PIK3CA-H1047R-wt) and mutant probe (PIK3CA-H1047R-mt), digested normal human gDNA and digested mutant plasmid.
[0035] The primers and probes are self-designed and optimized through multiple combinations, and the primers and probes include the homologous region of the mutant fragment inserted in the mutant plasmid. The primers and probes were synthesized by Shanghai Bailige Biotechnology Co., Ltd. The nucleotide sequences of the primers and probes are shown in Table 1.
[0036] Table 1 Primer Probe Sequence List
[0037] name
Sequence(5'—3')
Seq No.
PIK3CA-H1047R-F
GCAAGAGGCTTTGGAGTATTTCATG
1
PIK3CA-H1047R-R
GCTGTTTAATTGTGTGGAAGATCCAA
2
PIK3CA-H1047R-wt
CCACCATGATGTGCATC
3
PIK3CA-H1047R-mt
CACCATGACGTGCATC
4
[0038] T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com