Methods of producing a fermentation product in trichoderma
A fermentation product, Trichoderma reesei technology, applied in biochemical equipment and methods, fermentation, enzymes and other directions, can solve problems such as ineffective utilization of sucrose
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
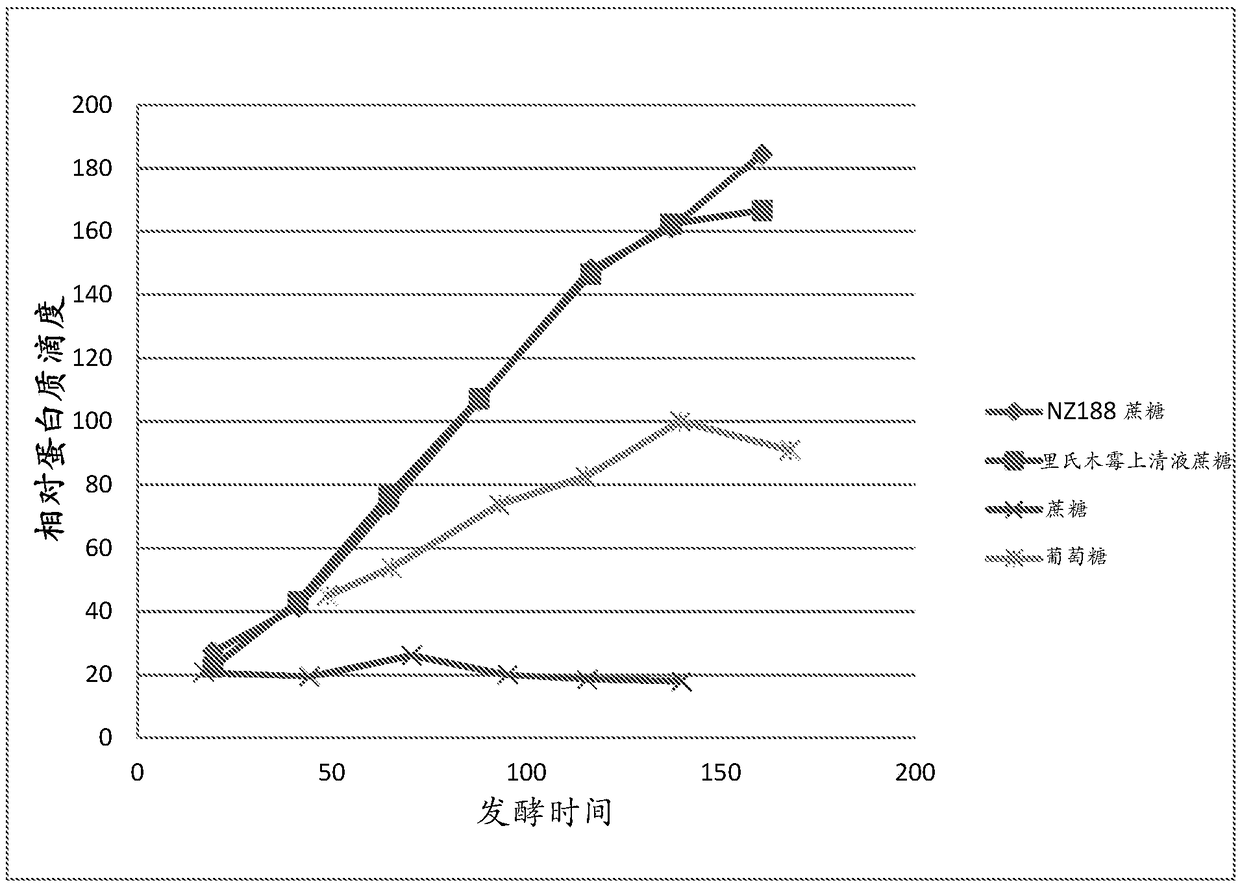
Image
Examples
example
[0073] Materials and Methods
[0074] Growth medium:
[0075] Fermentation medium
[0076]
[0077] Seed medium NNCell1:
[0078]
[0079]
[0080] SOC medium
[0082] 5g / L yeast extract
[0083] 4.8g / L MgSO 4
[0084] 3.603g / L dextrose
[0085] 0.5g / L NaCl
[0086] 0.186g / L KCl
[0087] 2XYT board
[0089] 10g / L yeast extract
[0090] 5g / L NaCl
[0092] COVE board
[0093] 342.3g sucrose
[0094] 20ml Cove's saline solution
[0095] 10ml 1M acetamide
[0096] 10ml 1.5M CsCl
[0097] 25g agar (Noble)
[0098] water to 1 liter
[0100] 26g KCl
[0101] 26g MgSO 4 7H 2 o
[0102] 76g KH 2 PO 4
[0103] 50ml COVE Trace Elements
[0104] water to 1 liter
[0105] COVE trace elements
[0106] 0.04g Nz 2 B 4 o 7 10H 2 o
[0107] 0.4g CuSO 4 5H 2 o
[0108] 1.2g FeSO 4 7H 2 o
[0109] 0.7g MnSO 4 h 2 o
[0...
example 1
[0133] Example 1 clone and prepare the plasmid encoding Aspergillus niger suc1 gene
[0134] The expression vector pMJ09 (WO 2005 / 067531) was used as the basis of the expression vector of this example.
[0135] The A. niger suc1 gene encoding invertase was PCR amplified from genomic DNA prepared from A. niger ATCC 1015 using the PCR primers shown below:
[0136] Suc1 F vector flk attacgaattgtttaaacgt gctttacttcactcgtgcatgggg (SEQ ID NO:2)
[0137] Suc1 R vector flk aaatggattgattgtct caccacgtgcacattcatattccgc (SEQ ID NO:3)
[0138] The underlined bases correspond to the gene sequence of Suc1, while the ununderlined bases correspond to the vector sequence.
[0139] reaction mixture
[0140]
[0141] PCR conditions:
[0142] Step 1 98°C for 30 seconds
[0143] Step 2 98°C for 10 seconds
[0144] Step 3 56°C for 15 seconds
[0145] Step 4 72°C for 160 seconds
[0146] Steps 2-4 were repeated for 34 cycles, and then the reaction mixture was kept at 10 °C.
[0147]...
example 2
[0152] Example 2 Transformation of Trichoderma reesei with pVCK12TRI001 comprising Aspergillus niger suc1 gene
[0153] Plasmid pVCK12TRI001 was linearized with the restriction endonuclease PmeI and transformed into Trichoderma reesei RutC30 strain essentially as described in Example 6 of WO 2008 / 151079, and the transformation was plated onto COVE plates.
[0154] Twenty one transformants were selected and transferred to plates of COVE2 + 10 mM uridine and incubated at 28°C for 22-26 days.
[0155] Transformants were subcultured onto new COVE2 plates, Trichoderma minimal plates + 2% sucrose and Trichoderma minimal plates + 2% glucose and incubated at 28°C for 8 days. All transformants grew well on Trichoderma minimal plates + 2% sucrose, Trichoderma minimal plates + 2% glucose and COVE2 plates. The untransformed Trichoderma reesei RutC30 strain did not grow on Trichoderma minimal plates + 2% sucrose, but grew as expected on Trichoderma minimal plates + 2% glucose.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com