Method, primers, probe and detection agent for tumor gene mutation detection
A mutation detection and tumor gene technology, applied in the field of molecular biology, can solve the problems of time-consuming, inability to detect and distinguish T790M/C797S cis and trans mutations, and high cost, so as to avoid human misjudgment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
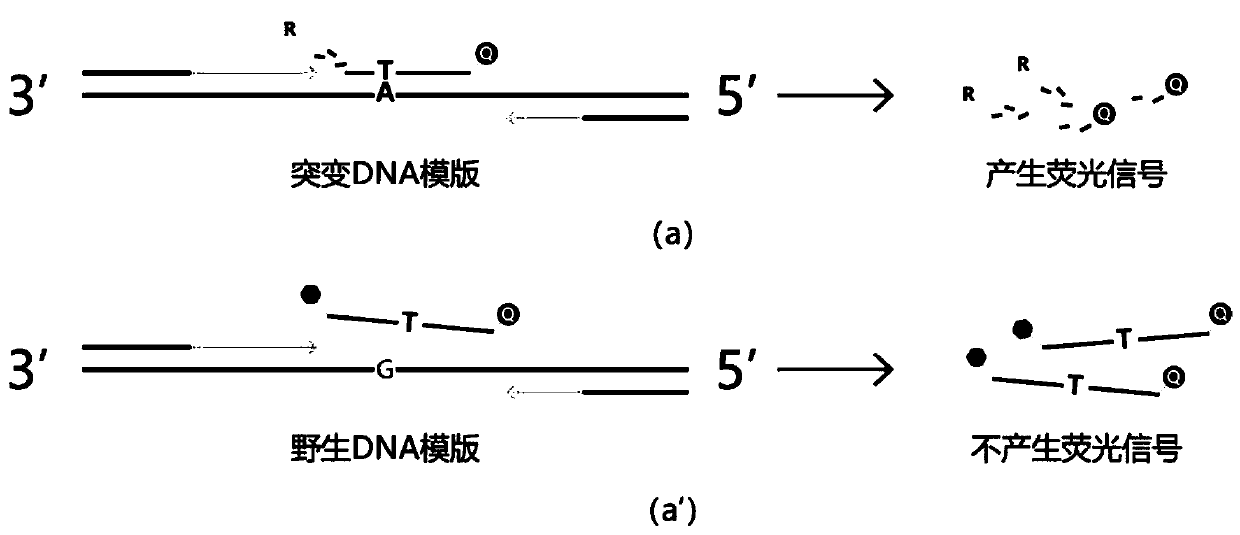
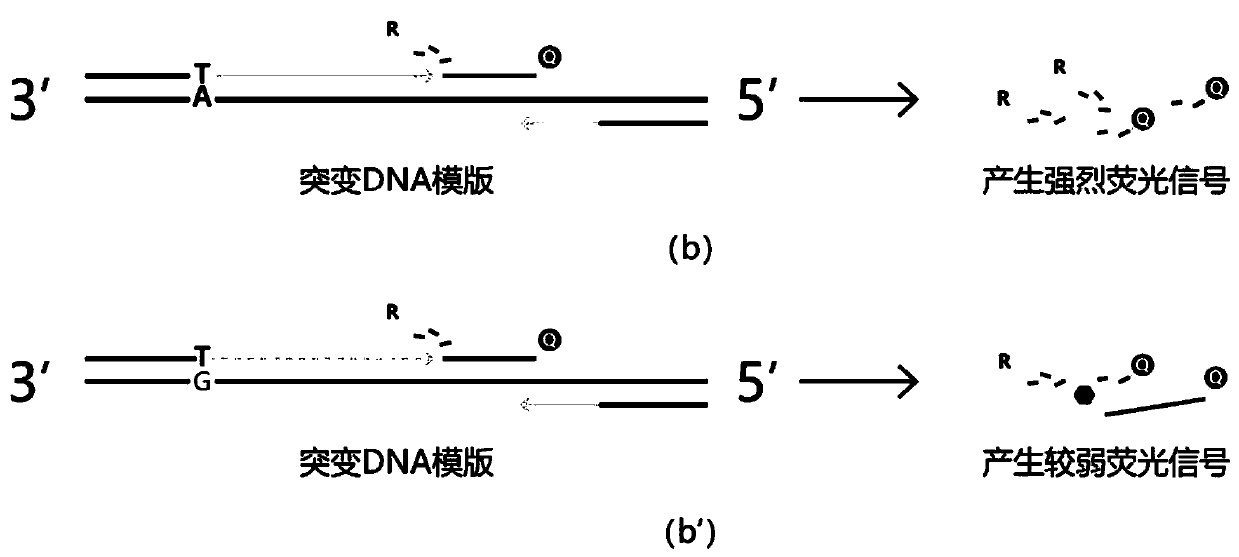
[0055] Detection is performed on the V600E mutation site of the BRAF gene, and the sample in Example 1 is a point mutation. The primer sequences used are as follows:
[0056]primer-F1: CTTCATGAAGACCTCACAGTAAAAATAGG
[0057] primer-R1: ACAAAATGGATCCAGACAACTGTTC
[0058] The probe sequences employed are as follows:
[0059] primer-P1: FAM-TCTAG GA ACAGAGAA CC - MGB.
[0060] The detection steps are as follows:
[0061] (1) Extraction of DNA / RNA from positive and wild-type tissue samples: AllPrep DNA / RNA / miRNA Universal Kit (QIAGEN Cat. No. 80224) was used for extraction, and the specific extraction steps were operated according to the kit instructions. The extracted DNA was diluted to 5ng / μL, and the RNA was diluted to 10ng / μL.
[0062] (2) The wild-type sample that has been determined to have no mutation is used as a negative control.
[0063] (3) Prepare PCR reaction solution according to the DNA PCR reaction system of Table 1 and the RNA PCR reaction system of Table...
Embodiment 2
[0082] The V769_D770insASV mutation site of the EGFR gene was detected. The sample mutation type in Example 2 is repetitive insertion of short segments. The primer sequences used are as follows:
[0083] primer-F2: GCCACACTGACGTGCCTCTC
[0084] primer-R2: GTCCAGGAGGCAGCCGAAGG
[0085] The probe sequences employed are as follows:
[0086] primer-P2: FAM-CCAGCG ACAG CAGCGTG-BHQ.
[0087] The detection steps refer to Example 1. The test results of the samples in Example 2 are consistent with the results of the first-generation sequencing.
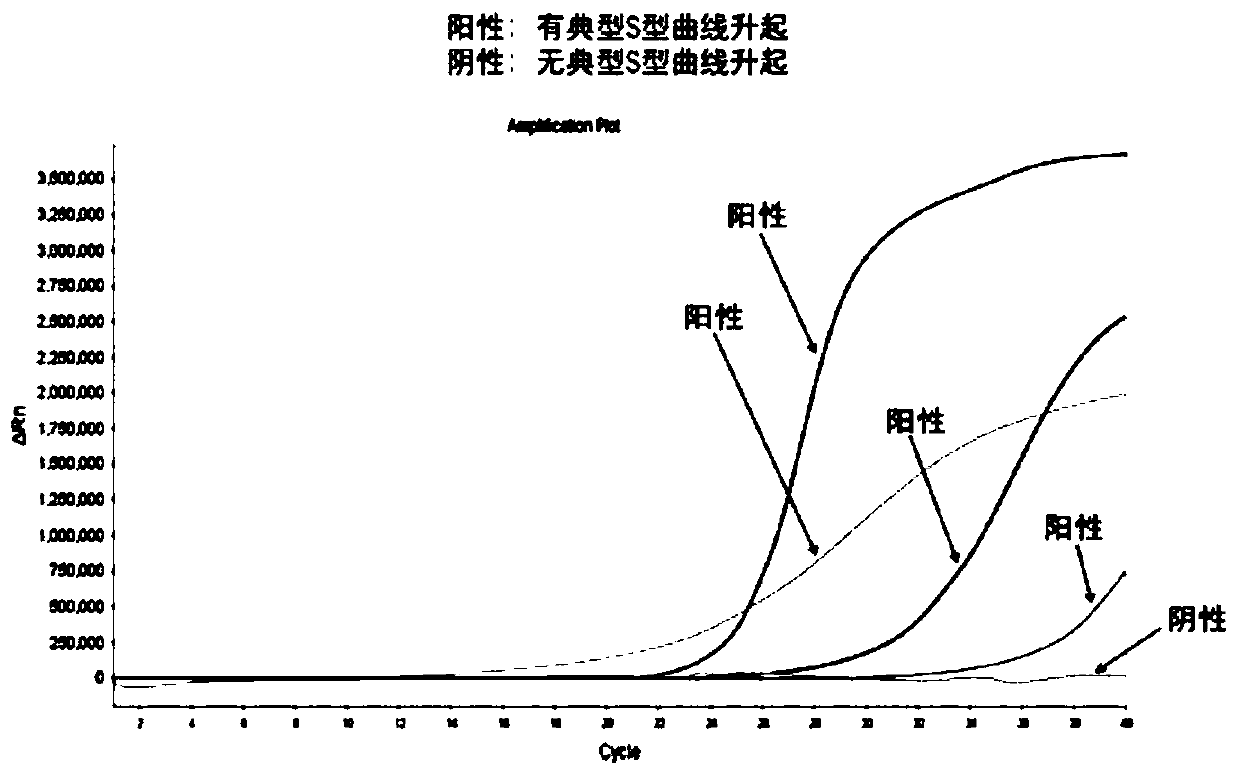
[0088] The detection sensitivity of embodiment 2 can reach 2.0% (see Figure 8 ).
Embodiment 3
[0090] Detection of the A775_G776insYVMA mutation site of the HER2 gene. The sample mutation type in Example 3 is repetitive insertion of short segments. The primer sequences used are as follows:
[0091] primer-F3: CGTGATGGC ATACGTGATGGC
[0092] primer-R3: CCAGCTGCACCGTGGATGTCAG
[0093] The probe sequences employed are as follows:
[0094] primer-P3: FAM-C TC TATGTCTCCCGCCTTCTG-BHQ.
[0095] The detection steps refer to Example 1. The test results of the samples in Example 3 are consistent with the results of the first-generation sequencing.
[0096] The detection sensitivity of embodiment 3 can reach 0.8% (see Figure 9 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com