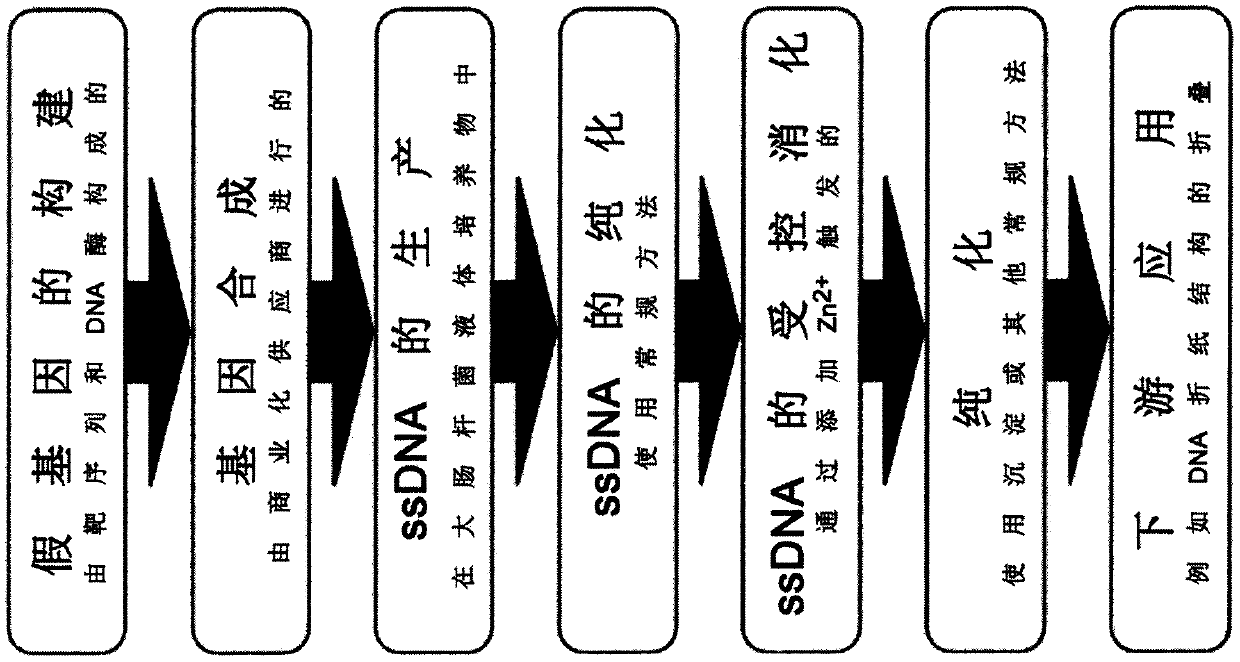
Scalable biotechnological production of DNA single strand molecules of defined sequence and length
A DNase and DNA sequence technology, which is applied in the field of recombinant production of DNA single-stranded molecules, can solve the problem that starting materials cannot be obtained in a large-scale manner
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0175] Example 1 Production of DNA-based nanorods
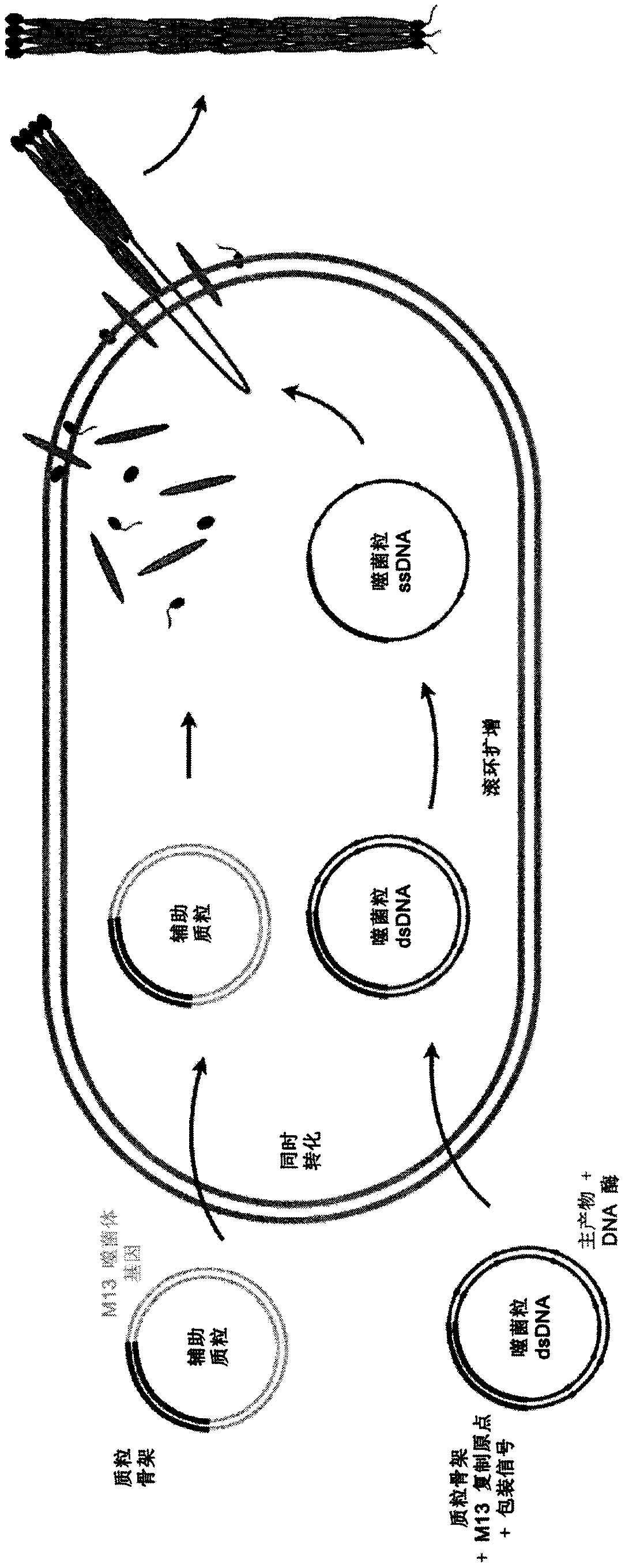
[0176] As a prototype, DNA-based nanorods were produced from DNA material produced exclusively by the method of the present invention. The DNA-based nanorods were designed and developed using the DNA origami design approach (see Rothemund, 2006; see also U.S. Patent Application Nos. 2007 / 117109 A1 and 2012 / 251583 A1; U.S. Patent Nos. 7,842,793 B2 and 8,501,923 B2). Ten DNA double helices arranged in parallel in a crystal lattice and cross-linked by strand junctions (see Figure 4 A). For the nanorods, a 2500 base long single-stranded DNA backbone molecule (the "scaffold strand") and 21 DNA oligonucleotides each approximately 100 bases in length (the "master product strand") are required . All required construction elements are derived from phagemids specially constructed for this purpose.
[0177] 1.1 Materials and methods
[0178] -sequence:
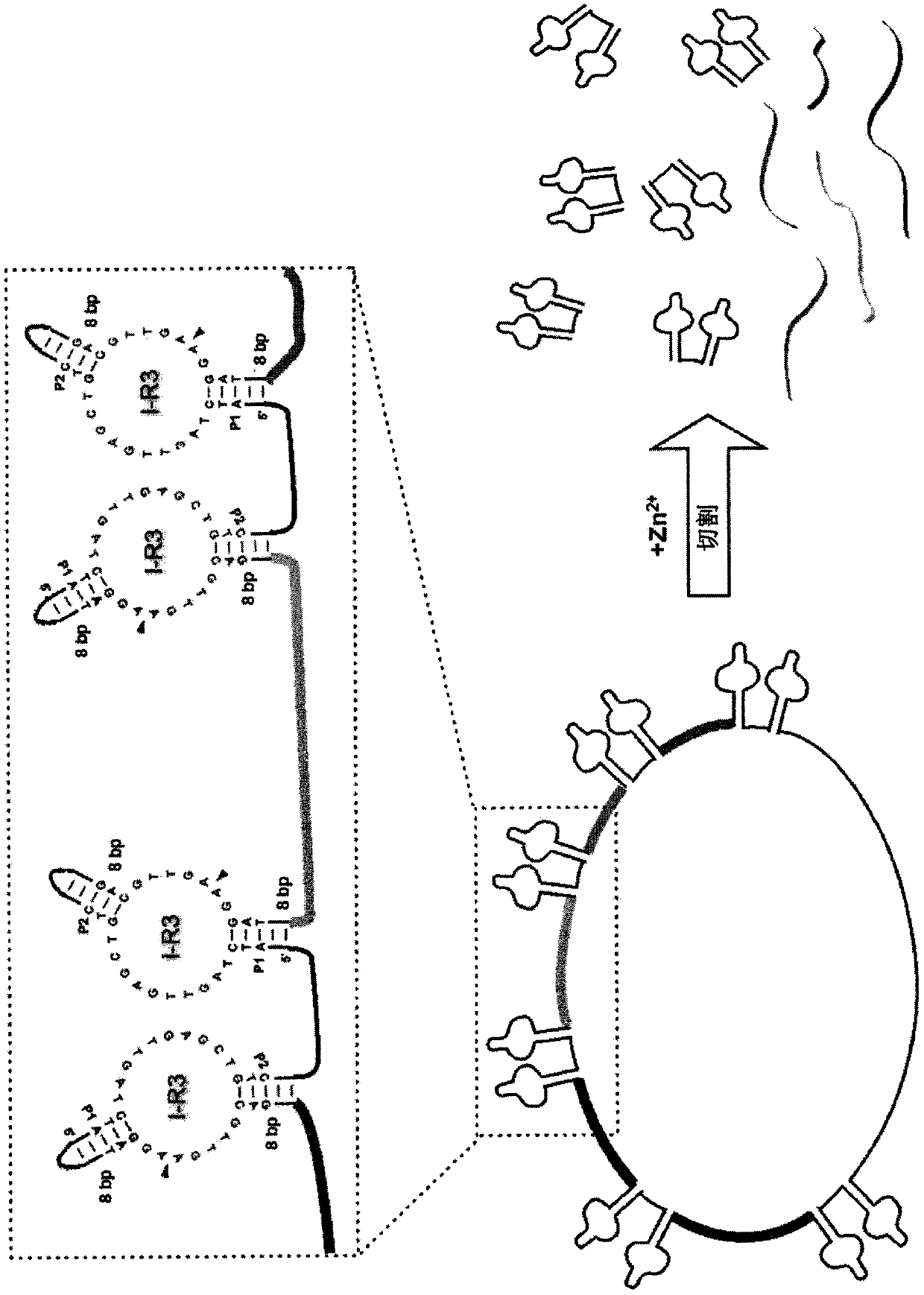
[0179] DNAse:
[0180] P1-[SEQ ID NO.1]-P2-P3-P2'-[SEQ ID NO.2]-P1'
[018...
Embodiment 2
[0200] Example 2 Optimization of DNase sequence
[0201] To construct our phagemids, we did not simply place a constant DNase sequence between the target oligonucleotides in the same manner as we inserted restriction enzyme binding sites. The sequence of the DNase depends on the terminal sequence of the oligonucleotide to be produced. Whereas Gu et al. (Biotechniques 2013) simply used the same end sequence and thus the same DNase, our method uses a different DNase sequence to produce different target oligonucleotide sequences.
[0202] To identify essential bases in the DNase sequence, we screened about 40 different variants of each DNase as oligonucleotides (two examples see Figure 6A ) and five versions of full-length phagemids (for two examples see Figure 6B ). We found some bases that were not classified as essential in the original literature on DNase (Gu et al., JACS 2013). This allows us to construct phagemids that are fully cleaved to the desired product in an ac...
Embodiment 3
[0220] Example 3 Other nanostructures assembled from DNA oligonucleotides produced using the DNase-based method of the invention
[0221] In addition to nanorods, we designed 48-helix tubes assembled from 3200 base-long scaffolds and 31 main product oligonucleotides (see Figure 7A ). As with nanorods, all major products for this 48-helix tube are encoded on one phagemid, and the backbone of the phagemid acts as a scaffold. As with the nanorods, due to the inherently precise 1:1 stoichiometry, the objects assembled efficiently into the desired shape without using the primary product in excess of the scaffold.
[0222] To demonstrate the applicability of our method to the assembly of existing full-scale DNA origami objects, we generated all 161 Phagemids seeded with primary product oligonucleotides (see Figure 7B ). In contrast to nanorods and 48-helix bundles, the main product for pointers is distributed on 4 separate phagemids and uses a single scaffold (M13mp18, 7249 ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com