Protein proximity labeling method based on DNA nanotechnology and application thereof
A labeling method and nanotechnology technology, applied in the field of adjacent labeling, can solve the problems of affecting the function of the target protein, locating interactomics, affecting the accuracy and specificity of mass spectrometry identification, and complex operation, so as to improve the labeling efficiency and labeling efficiency. High and specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1 A protein proximity labeling method based on DNA nanotechnology, comprising the following steps:
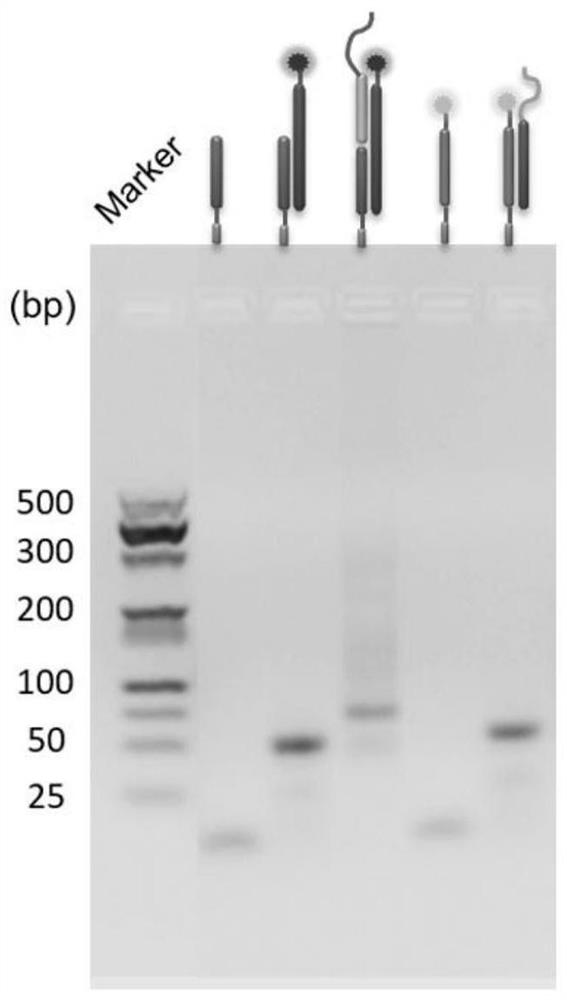
[0034] (1) Synthesis of DNA nanostructures: The designed DNA sequences S1 (SEQ ID NO:1), S3 (SEQ ID NO:3) and S4 (SEQ ID NO:4) were mixed in TE buffer (10mmol / LTris- HCl; 1 mmol / L EDTA; pH 7.8-8.2), the DNA sequences S2 (SEQ ID NO: 4) and S5 (SEQ ID NO: 5) were mixed in equal proportions in TE buffer (10 mmol / LTris-HCl; 1 mmol at the same time) / L EDTA; pH 7.8-8.2), then put the two sequences into the PCR synthesizer respectively, annealed at 95°C for 10 minutes, then rapidly cooled from 95°C to 4°C, and finally kept at 4°C, and finally the single-stranded S1, S3 and S4 synthesize DNA nanostructure A, and single-stranded S2 and S5 synthesize DNA nanostructure B. Details of DNA sequences S1-S5 are shown in Table 1:
[0035]Table 1 DNA sequences
[0036]
[0037] During the synthesis of DNA nanostructures, agarose gel electrophoresis was used to verify whe...
Embodiment 2
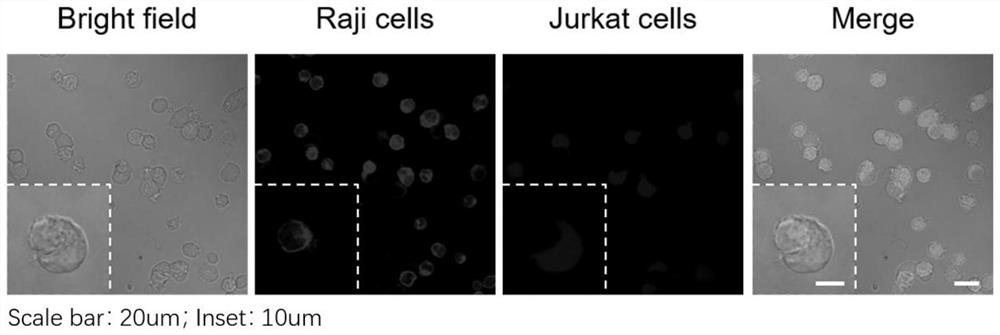
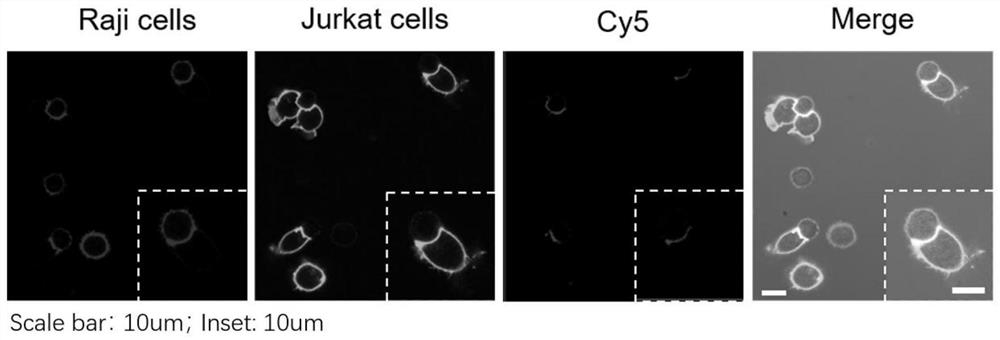
[0044] Example 2 A protein proximity labeling method based on DNA nanotechnology and its immunoblotting and mass spectrometry identification
[0045] The synthetic steps of step (1)-(4) are identical with embodiment 1;
[0046] (5) Lyse cells to extract membrane proteins: use Mem-PER TM The Plus Membrane Protein Extraction Kit (purchased from ThermoFisher Scientific, Cat. No. 89842) was used to extract membrane proteins. Specifically, 1 mL of permeabilization buffer was added, and the cells were lysed at 4°C for 10 minutes. After centrifugation to remove the supernatant, 500 uL was added to the pellet for solubilization. buffer, incubated at 4°C for 30 minutes, and the supernatant obtained after centrifugation was the cell membrane protein.
[0047] (6) Enrichment of target protein: The target protein labeled with biotin was enriched with streptavidin-coupled magnetic beads (purchased from BeaverBio, Item No. 22307).
[0048] (7) Western blotting: add 20uL of 1× protein loa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com