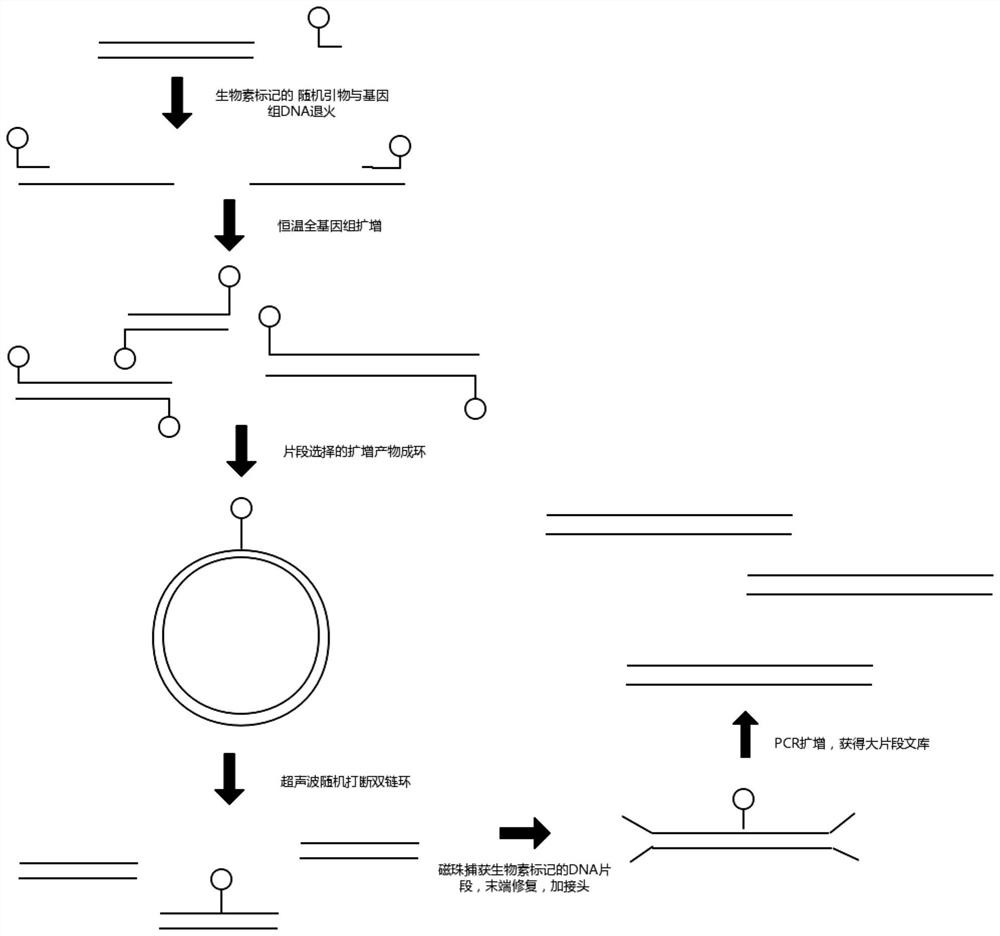
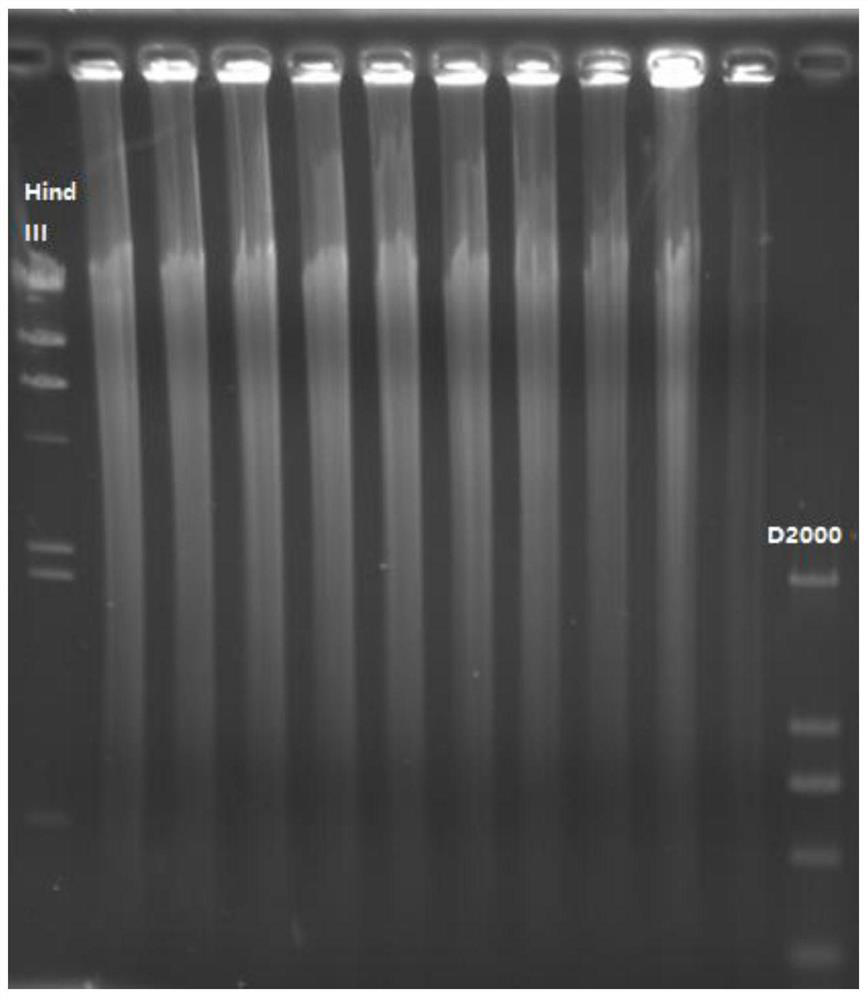
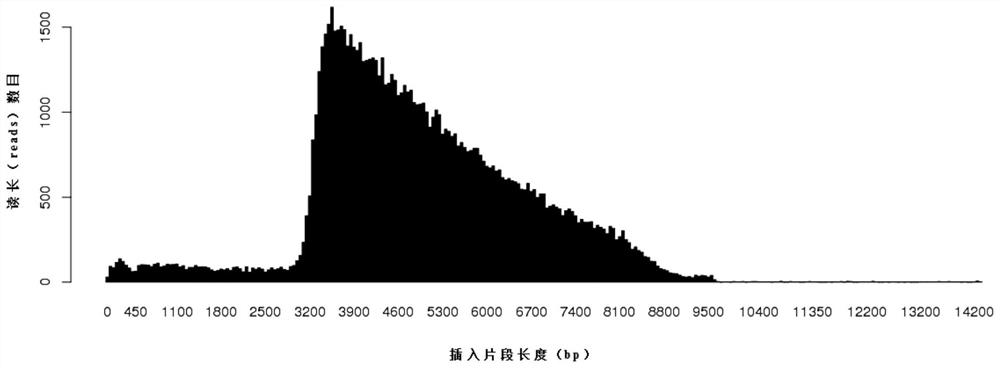
dna library construction method with low input
A DNA library and construction method technology, which is applied in DNA preparation, recombinant DNA technology, chemical library, etc., can solve the problems that samples are difficult to obtain and cannot meet the needs of large fragment library construction, and achieve the effect of reducing the experimental process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0039] The present invention will be further described in detail below through specific embodiments in conjunction with the accompanying drawings. In the following implementation manners, many details are described for better understanding of the present application. However, those skilled in the art can easily recognize that some of the features can be omitted or replaced by other materials and methods in different situations.
[0040]In addition, the characteristics, operations or characteristics described in the specification can be combined in any appropriate manner to form various embodiments. At the same time, the steps or actions in the method description can also be exchanged or adjusted in a manner obvious to those skilled in the art. Therefore, various sequences in the specification and drawings are only for clearly describing a certain embodiment, and do not mean a necessary sequence, unless otherwise stated that a certain sequence must be followed.
[0041] The m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com