The Molecular Marker brsf0239 Primer and Its Application of the Major Qtl Locus in Flowering and Ripening Stages of Brassica napus
A Brassica napus and molecular marker technology, which is applied in the fields of molecular biology and genetic breeding, can solve the problems of difficult rapeseed breeding application, small repeatability, high technical difficulty, etc., and achieves convenient, fast, accurate and rapid detection methods, and improved selection. The effect of efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Construction and trait determination of segregating populations at flowering stage of rapeseed:
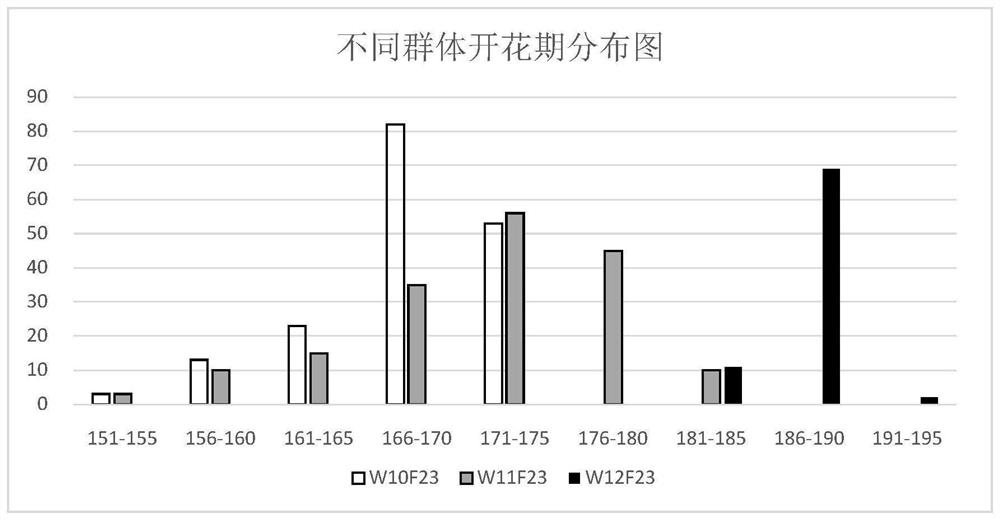
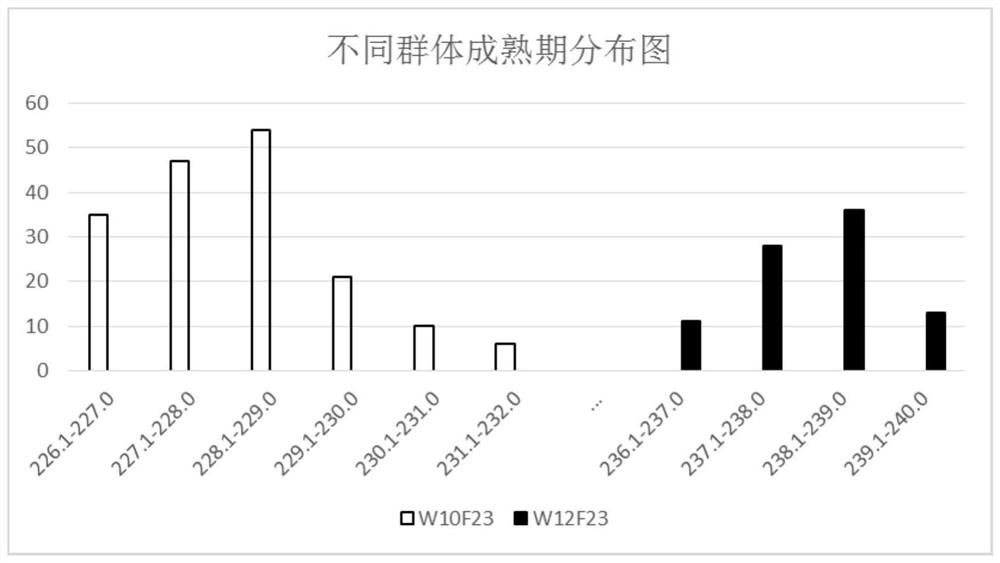
[0028] In this example, Shuang11, a rapeseed variety sequenced, was used to cross with another rapeseed variety 73290 with large genetic differences, and the F1 generation of the hybrid was selfed to generate the F2 population and its F2:3. Among them, the Yangluo Comprehensive Experimental Base of the Institute of Oil Plants, Chinese Academy of Agricultural Sciences, planted F2 populations in 2009, and planted F2:3 family populations in 2010, 2011 and 2012. The anthesis and maturity phenotypes of the two parents and segregating populations were identified after harvest at maturity. The test data showed that the mean values of flowering stage and maturity stage in the three environments were normally distributed, indicating that the quantitative genetic characteristics of flowering stage and maturity stage traits ( figure 1 , figure 2 ).
Embodiment 2
[0030] Primer development and synthesis:
[0031] The SSR primers used by the applicant include two types: one is the published primer sequence in published articles and Brassica database (http: / / www.brassica.info / resource / markers / ssr-exchange.php); the other is It was developed by the applicant based on the sequences of cabbage and cabbage scaffolds, named BrSF and BoSF series respectively. The specific development method is to use SSR Hunter software to search for SSRs in each scaffold, and then use Primer3.0 software to design SSR primers. The self-developed InDe l primers are obtained by comparing the resequencing sequence of 73290 with the reference genome sequence of Zhongshuang11. First, the resequencing sequence of 73290 is positioned on the whole genome reference sequence of Zhongshuang11 by using BW A software , and then use samtools software to find InDel.
Embodiment 3
[0033] The screening process of primer polymorphism is as follows:
[0034] (1) Randomly select 10 strains of DNA from each parent and mix them in equal amounts as templates for screening primers.
[0035] (2) Utilize the primers after dissolving to carry out PCR amplification to the parental DNA,
[0036] reaction system:
[0037]
[0038] PCR reaction program:
[0039]
[0040]
[0041] (3) Gel electrophoresis band pattern interpretation
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com