Long-chain non-coding RNA subcellular localization method based on multi-feature information fusion
A technology for long-chain non-coding and subcellular localization, applied in the new field of long-chain non-coding RNA subcellular localization, can solve the problem of inaccurate prediction of subcellular location
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0048] The present invention will be described in further detail below in conjunction with the accompanying drawings.
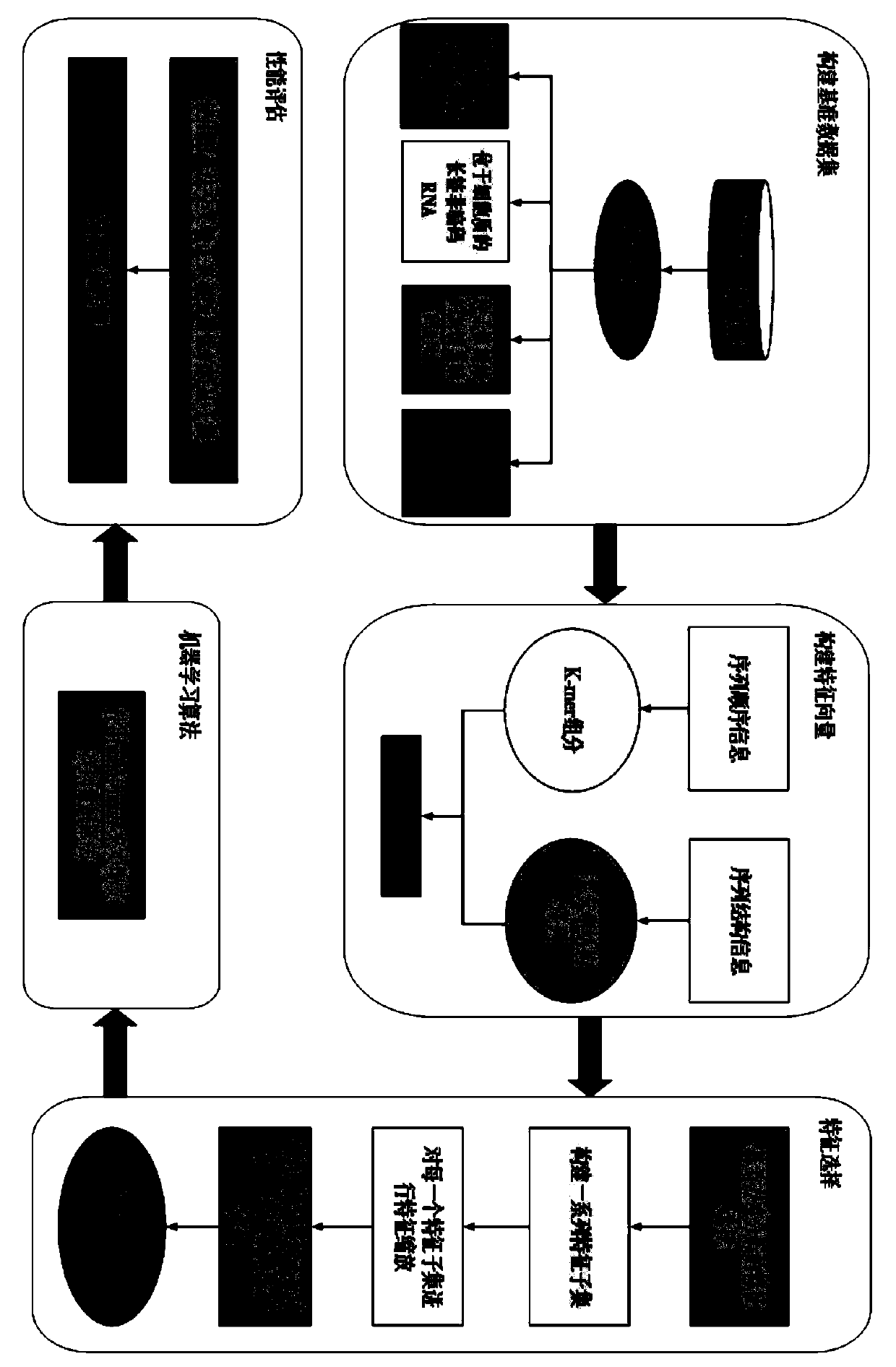
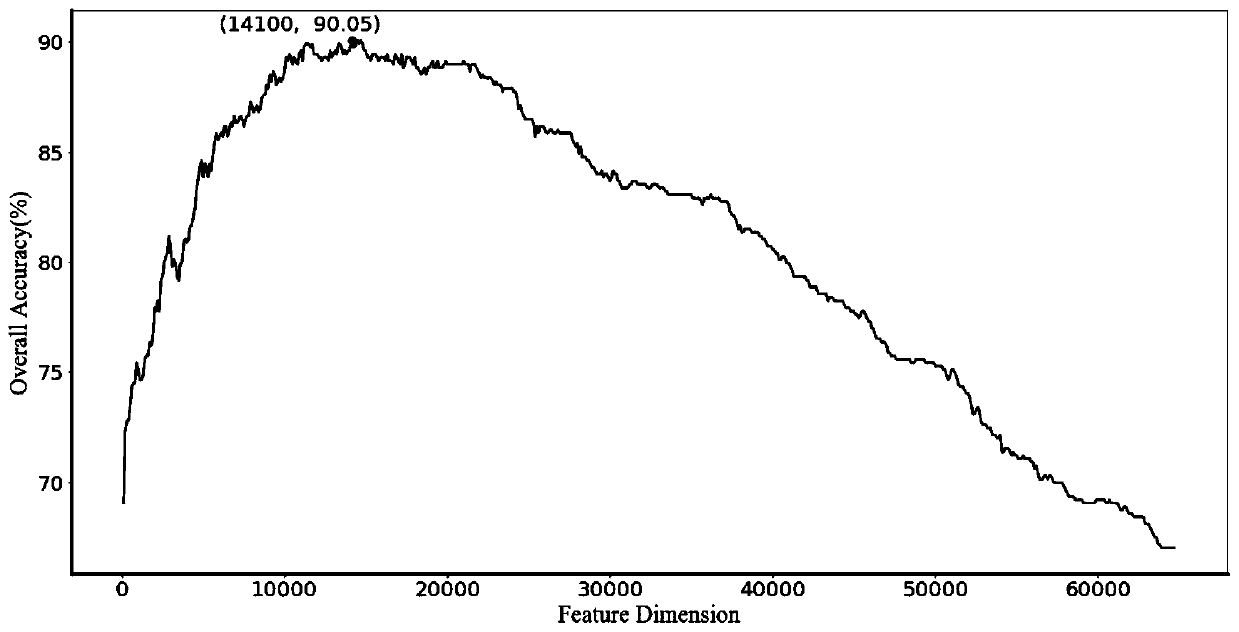
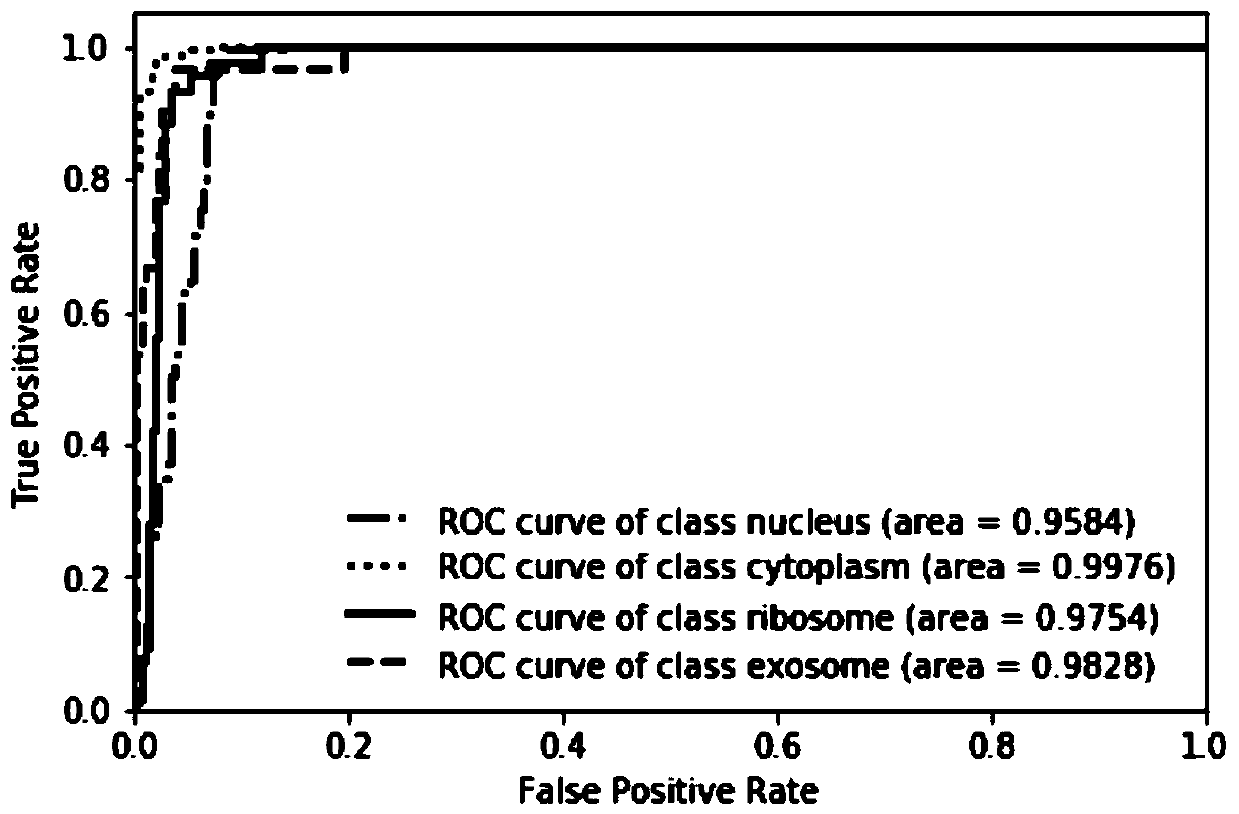
[0049] see figure 1 , the present invention mainly includes 5 parts, (i) construct benchmark data set. By screening the data in the RNALocate database, 643 long non-coding RNA sequences located in different subcellular locations were obtained. (ii) Construct feature vectors. By fusing the k-mer components of long-chain non-coding RNAs with the triplet structure-sequence to form feature vectors, the sequence and structure information of long-chain non-coding RNAs is more comprehensively utilized. Since the 8-mer component has a unique evolutionary mechanism, the parameter k is set to 8, so far, we can express a long non-coding RNA sequence as (4 8 +32) dimensional feature vector. (iii) Feature selection. The method of analysis of variance is used to select the optimal feature subset. (iv) Apply machine learning algorithms. Choose a support vector machi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com