Gene sequencing data compression method, system and computer readable medium
A gene sequencing and data compression technology, applied in computing, electrical digital data processing, special data processing applications, etc., can solve the problems of different compression algorithm performance, long algorithm compression/decompression time, and reduced compression rate, and achieve short compression time. , the effect of stable compression performance and low compression rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
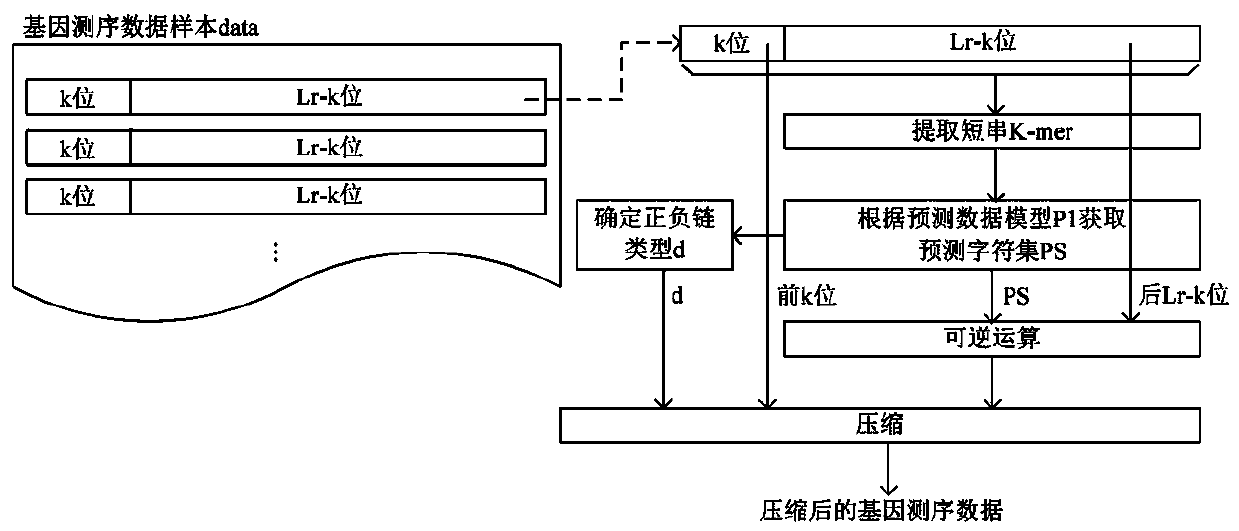
[0044] see figure 1 The method for compressing gene sequencing data in this embodiment includes:
[0045] 1) Traverse and obtain the read sequence R with the read length Lr from the gene sequencing data sample data;
[0046] 2) For each read sequence R, select k original gene letters as the original gene string CS 0 , from the original gene string CS 0 Start to generate a fixed-length k-bit string as a short string K-mer in a sliding window sequence of length k, determine the positive and negative strand type d of the read sequence R according to the short string K-mer, and use the preset prediction data model P1 Obtain the predicted character c corresponding to the adjacent bit of each short string K-mer to obtain a predicted character set PS with a length of Lr-k bits, and the predicted data model P1 includes any short string K- in the positive and negative strands of the reference genome mer and the predicted character c corresponding to its adjacent bits; the Lr-k origi...
Embodiment 2
[0091] This embodiment is basically the same as Embodiment 1, and the main difference is that the prediction data model P1 in step 1) is different.
[0092] In this embodiment, the prediction data model P1 is based on the base letter c corresponding to the short string K-mer in the reference genome and its adjacent bits in advance. 0 Complete the trained neural network model; Step 2.2.2) For each tuple (k-mer, 0) in the positive chain prediction sequence KP1, obtain its corresponding prediction character c through the mapping function mapping of the prediction data model P1. Input each tuple (k-mer, 0) in the positive chain prediction sequence KP1 into the neural network model to obtain the predicted character c corresponding to the tuple (k-mer, 0); step 2.2.4) for negative chain prediction Each tuple (k-mer, 1) in the sequence KP2 is mapped through the mapping function of the predicted data model P1 to obtain the predicted character c corresponding to its adjacent bits. Spec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com