Single-domain antibody for recognizing complex formed by HLA-A2 molecule and ITDQVPFSV short peptide
An HLA-A2, single-domain antibody technology, applied in the field of single-domain antibodies, can solve the problems of TCR mismatch, poor progress, difficult TCR acquisition, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
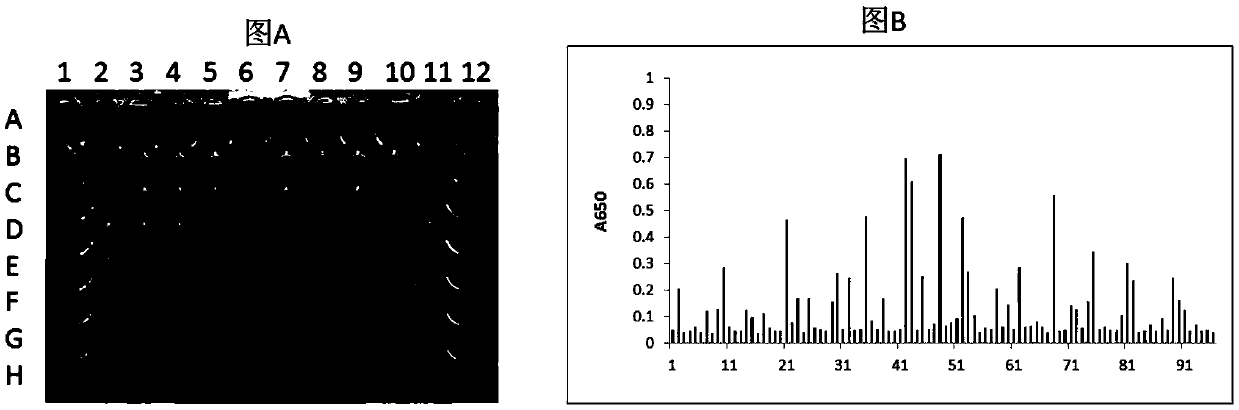
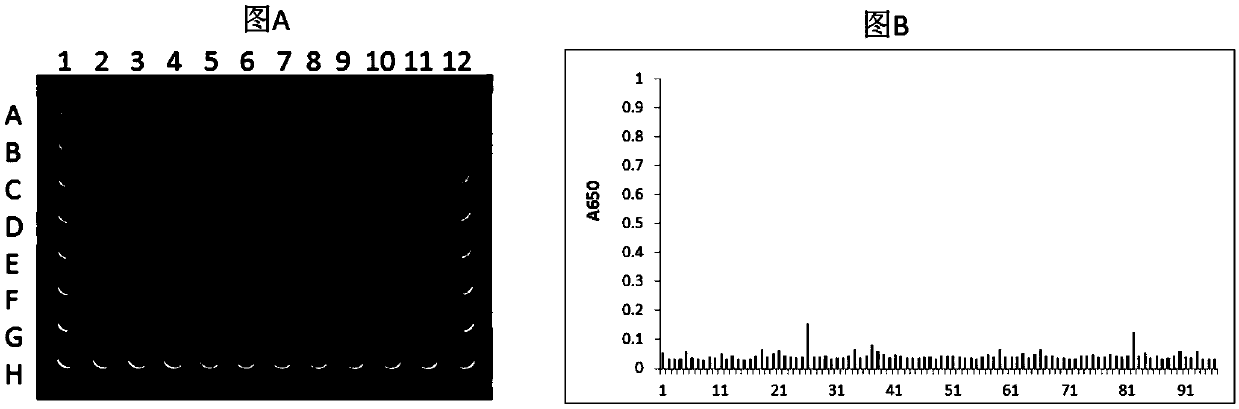
[0133] Example 1 Screening of single domain antibodies of HLA-A2 / ITDQVPFSV complex
[0134] 1.1 Preparation of single domain antibody phage library
[0135] 1.1.1 Preparation of helper phage (BM13)
[0136] The M13KE phage (purchased from NEB#N0316S) replicon was digested with AlwnI (purchased from NEB) and AfeI (purchased from NEB), and the artificially synthesized gene fragment was also digested with AlwnI and AfeI (purchased from NEB), and then Connect them with T4 ligase. After ligation, TG1 was transfected to obtain helper phage BM13. In this way, the original replicon tctggtggtggttctggtggcggctctgagggtggtggctctgagggtggcggttctgagggtggcggctctgagggaggcggttccggtggtggctct sequence is replaced by a synthetic gene sequence, that is, a trypsin cleavage sequence is added to the GIII coding region of the phage, which reduces the number of trypsin cleavage sequences that are used as a helper phage to digest the phage. Number of.
[0137] The synthetic gene sequence is as follows: CCA GCC...
Embodiment 2
[0167] Example 2. Preparation of fusion proteins M3-B2-Fc, M3-D9-Fc, M3-E7-Fc
[0168] 1. Construction of recombinant vector
[0169] The DNA molecule shown in SEQ ID No. 16 (which includes Kozak sequence and signal peptide, M3-B2 antibody sequence, human Fc sequence and tag sequence) was cloned into the HindⅢ and XbaI restriction endonuclease sites of pcDNA3.1 In time, the recombinant vector pcDNA3.1-M3-B2-Fc was obtained.
[0170] The DNA molecule shown in SEQ ID No.17 (which includes Kozak sequence and signal peptide, M3-D9 antibody sequence, human Fc sequence and tag sequence) was cloned into the HindⅢ and XbaI restriction enzyme sites of pcDNA3.1 In time, the recombinant vector pcDNA3.1-M3-D9-Fc was obtained.
[0171] The DNA molecule shown in SEQ ID No. 18 (including Kozak sequence and signal peptide, M3-E7 antibody sequence, human Fc sequence and tag sequence) was cloned into the HindⅢ and XbaI restriction endonuclease sites of pcDNA3.1 In between, the recombinant vector pcDN...
Embodiment 3
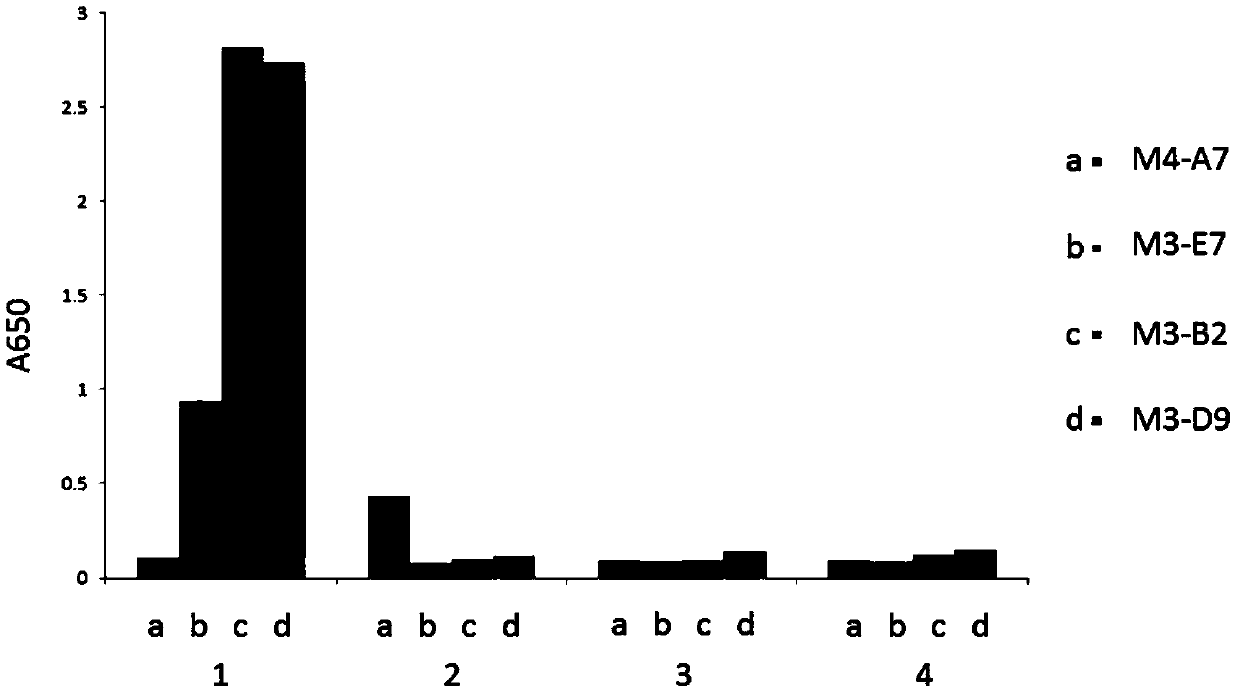
[0178] Example 3. Fusion proteins M3-B2-Fc, M3-D9-Fc, and M3-E7-Fc specifically recognize the HLA-A2 / ITDQVPFSV complex.
[0179] The fusion proteins M3-B2-Fc, M3-D9-Fc, M3-E7-Fc, and M4-A7-Fc after purification of Protein A were identified by ELISA. The specific steps of ELISA identification are as follows: Dilute the known antigen to 1-10μg / ml with coating buffer, add 0.1ml per well, overnight at 4℃; wash 3 times the next day; add 0.1ml of a certain diluted sample to be tested In the above-mentioned reaction wells coated with antigen, incubate at 37°C for 1 hour and wash; add 0.1ml of freshly diluted enzyme-labeled secondary antibody (horseradish peroxidase HRP-labeled anti-human antibody, 1:5000), Incubate at 37°C for 30 minutes and wash; wash with DDW for the last time. Add 0.1ml of the temporarily prepared TMB substrate solution to each reaction well, and let it stand at 37°C for 10-30 minutes. Use an advanced plate reader to read the plate at 650nm wavelength.
[0180] The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com