Nuclear localization signal F4NLS and application thereof in improvement of basic group editing efficiency and expansion of editable basic group range
A nuclear localization signal and encoding technology, applied in the biological field, can solve problems such as inability to be edited by bases, low base editing efficiency, and inability to be edited
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0213] Example 1. Application of the nuclear localization signal F4NLS in improving the efficiency of A·G base substitution or expanding the range of editable A
[0214] 1. Design and Construction of Recombinant Expression Vectors
[0215] 1. Design of recombinant expression vector
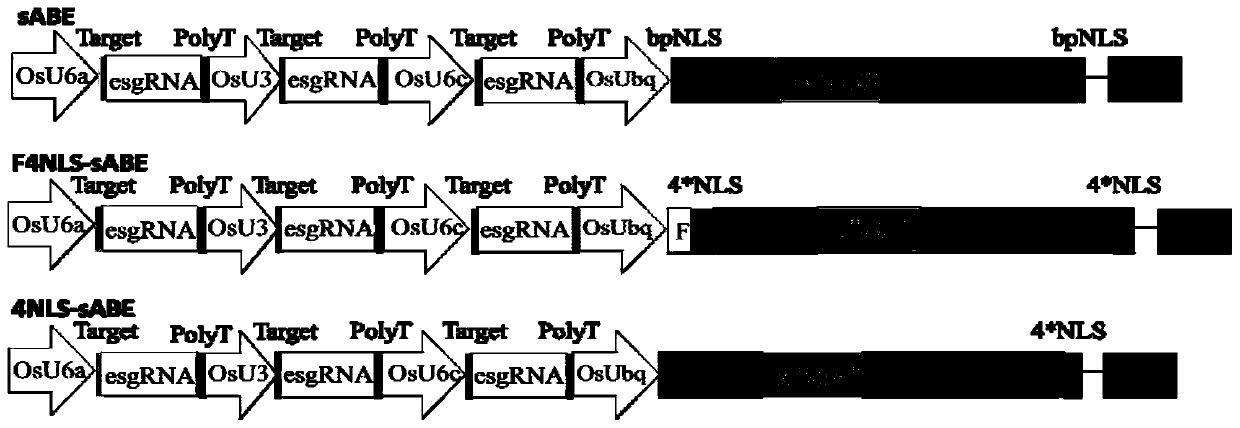
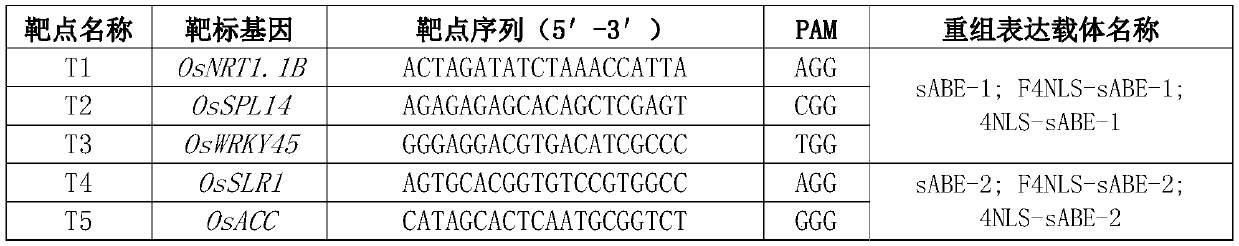
[0216] The nuclear localization signal is added to the ecTadA&ecTadA*&Cas9n base editing system. According to the added nuclear localization signal, it is divided into three design types, and the structure diagram of the recombinant expression vector containing three different design types is as follows: figure 1 shown. The design methods for the three different design types are as follows:
[0217] sABE system: add the bpNLS nuclear localization signal before the ecTadA element in the ecTadA&ecTadA*&Cas9n base editing system, and add the bpNLS nuclear localization signal after the Cas9n element. This design type is denoted as bpNLS-bpNLS. The amino acid sequence of the bpNLS nuclear localizat...
Embodiment 2
[0253] Example 2, the application of the nuclear localization signal F4NLS in improving the efficiency of C T base substitution
[0254] 1. Design and Construction of Recombinant Expression Vectors
[0255] 1. Design of recombinant expression vector
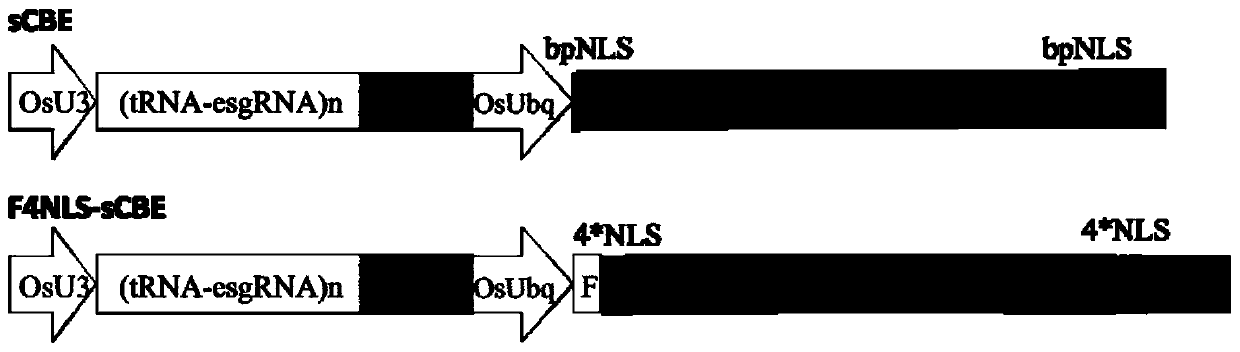
[0256] The nuclear localization signal is added to the rAPOBEC1&Cas9n&UGI base editing system. According to the added nuclear localization signal, it is divided into two design types. The structure diagram of the recombinant expression vector containing two different design types is as follows: figure 2 shown. The design methods for the two different design types are as follows:
[0257] sCBE system: add the bpNLS nuclear localization signal before the rAPOBEC1 element in the rAPOBEC1&Cas9n&UGI base editing system, and add the bpNLS nuclear localization signal after the UGI element.
[0258] F4NLS-sCBE system: Add 3*Flag&4*NLS nuclear localization signal before the rAPOBEC1 element in the rAPOBEC1&Cas9n&UGI base editing syste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com