CRISPR technology-based method for site-directed mutation of E. coli genes
A technology of Escherichia coli and recombinant Escherichia coli, applied in the biological field, can solve the problems of cumbersome process, low recombination efficiency and high off-target rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
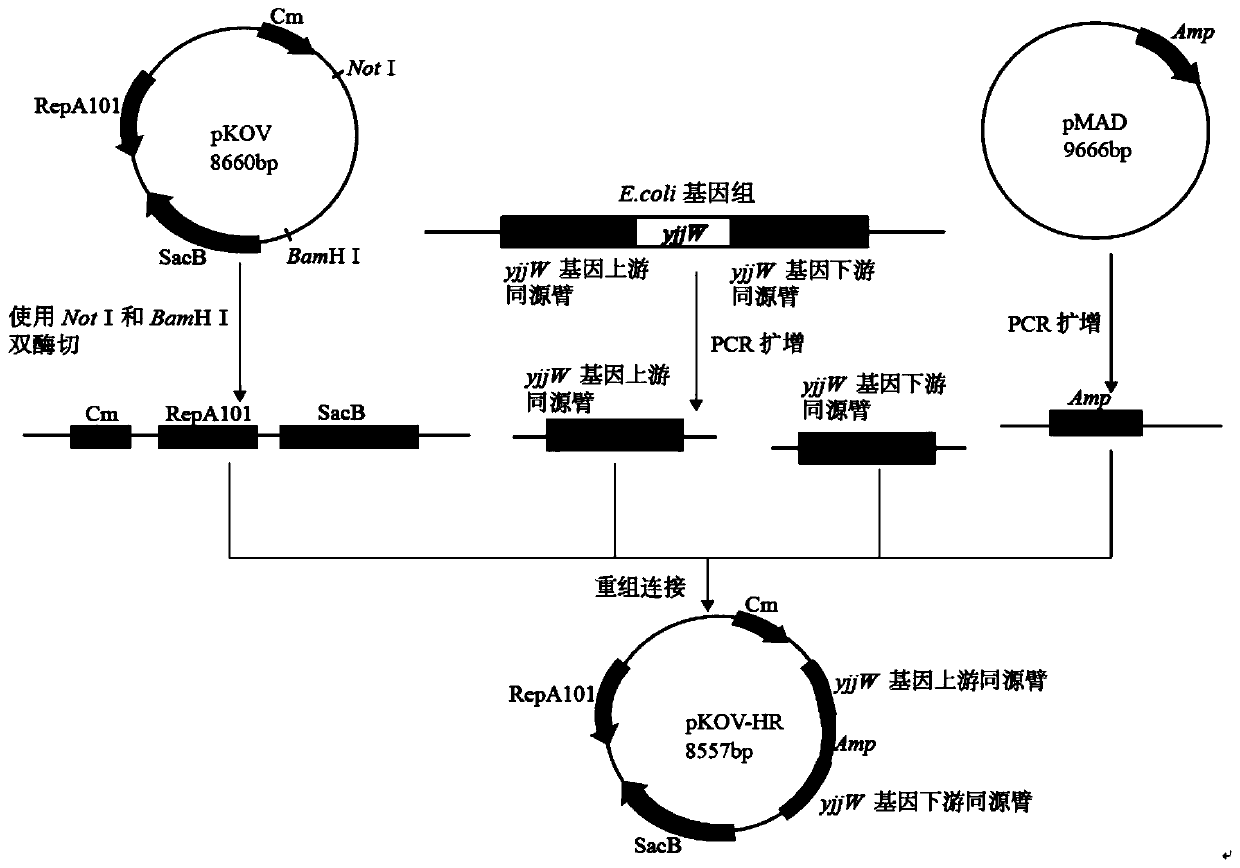
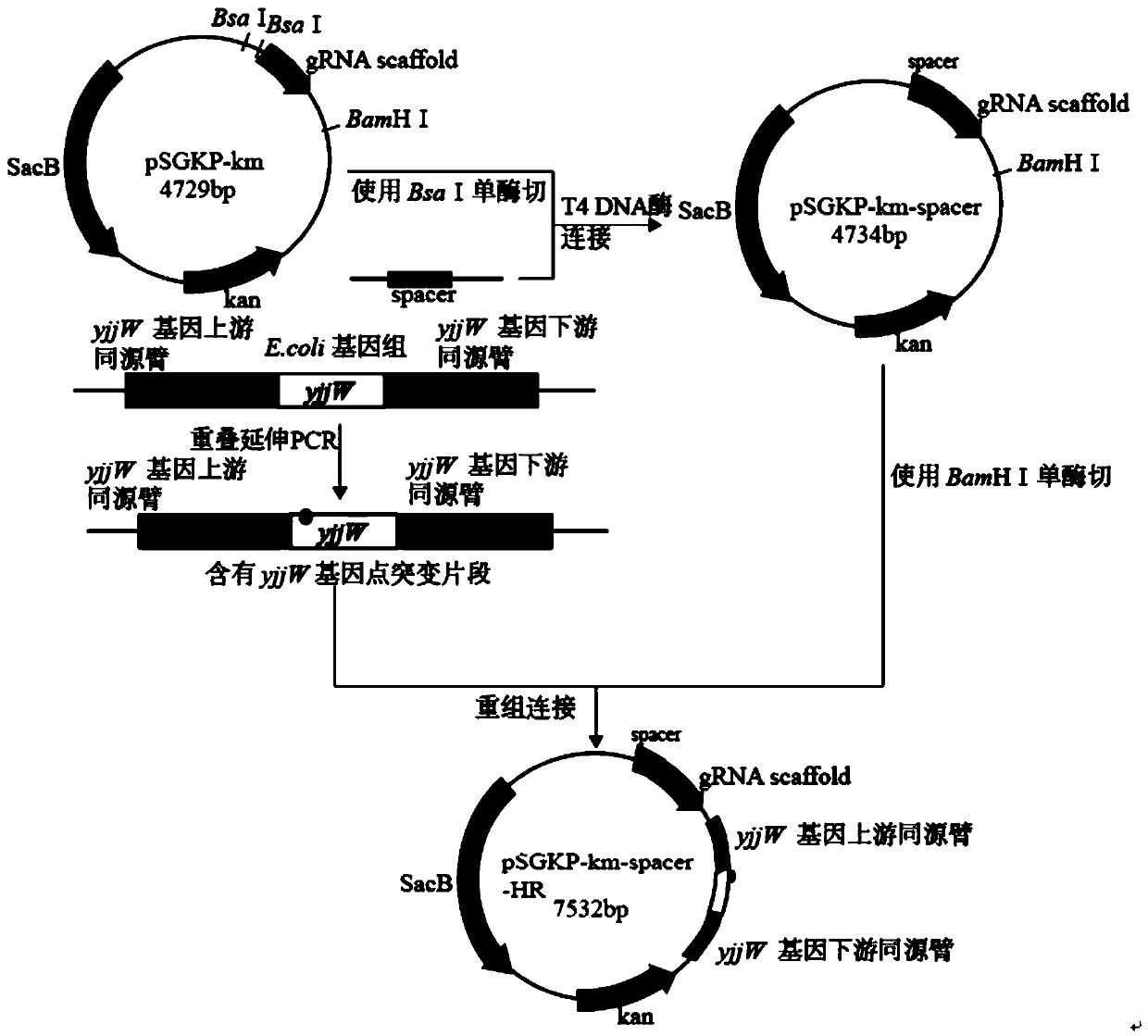
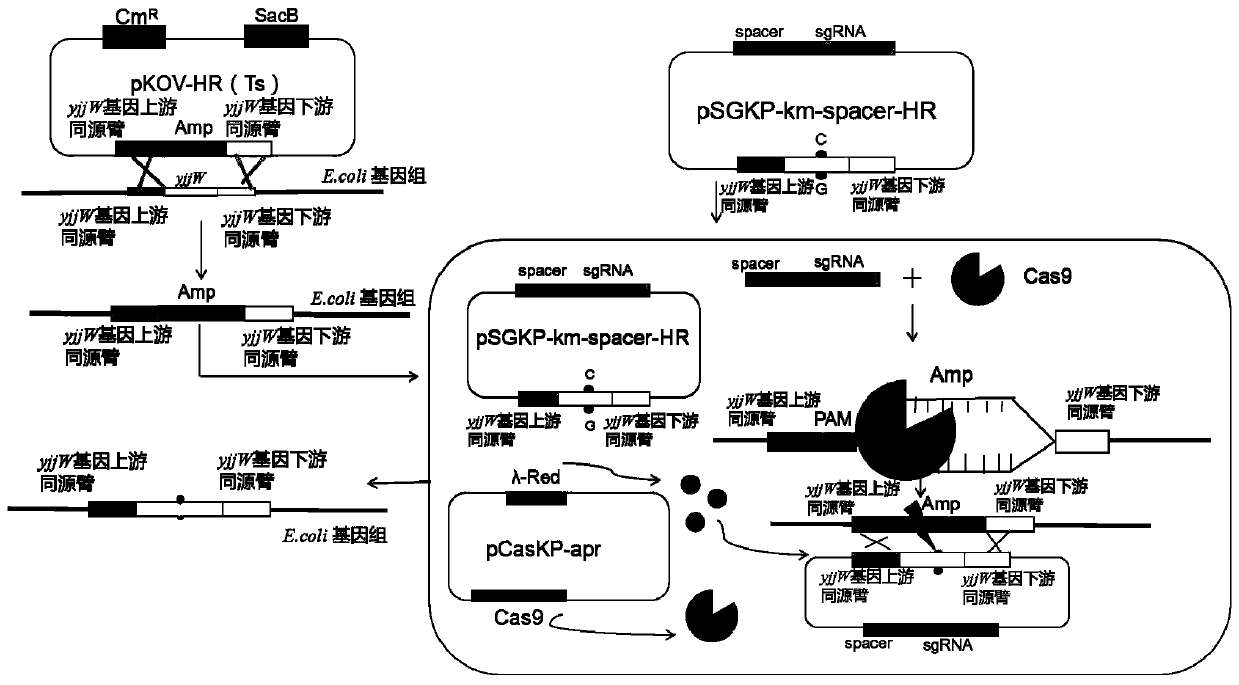
[0041] Example 1. Two-step legal point mutation of the target gene in the Escherichia coli genome based on CRISPR technology
[0042] 1. Materials
[0043] 1. Strains and plasmids
[0044]The strains and plasmid details used in the present invention are shown in Table 1.
[0045] Table 1 bacterial strain and plasmid used in the present invention
[0046]
[0047] [1] Wang Y, Wang S, Chen W, Song L-q, Zhang Y, Shen Z, Yu F, Li M, JiQ. Precise and efficient genome editing in Klebsiella pneumoniae using CRISPR-Cas9 and CRISPR-assisted cytidine deaminase. Applied and environmentalmicrobiology 2018,84(23):e01834-18.pSGKP-km and pCasKP-apr are available to the public from the applicant, and can be used to repeat the experiment of the present invention, and should not be used for other purposes.
[0048] 2. Primers
[0049] The primers used in the present invention are listed in Table 2.
[0050] Primers used in the present invention in table 2
[0051]
[0052] Note: The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com