Method for site-directed mutation of rice CENH3 gene by using CRISPR-CAS9 technology
A gene site-directed mutation, CRI-R2 technology, applied in recombinant DNA technology, horticultural methods, genetic engineering, etc., can solve the problems of cytotoxicity, cumbersome process, high off-target rate, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
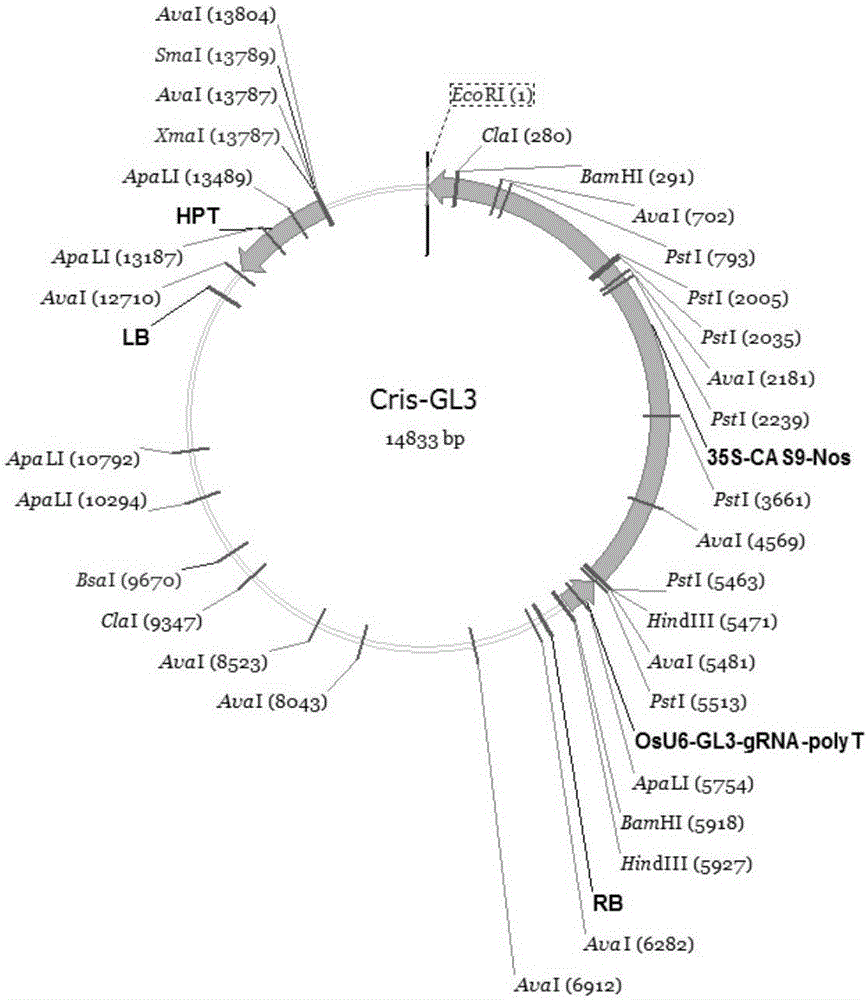
[0072] Example 1 Construction of carrier pHZ1-CAS9-gRNA-CENH3
[0073] 1. Selection of target sites in rice CENH3 (LOC_Os05g41080) gene
[0074] Log in to the plant transcription factor database (http: / / rice.plantbiology.msu.edu / analyses_search_locus.shtml), query the sequence of the rice CENH3 gene (LOC number: LOC_Os05g41080), and design the sgRNA sequence based on CRISPR / Cas9.
[0075] The nucleotide sequence of the sgRNA action site is 5'-GTCCAGGCACAGTGGCACTG-3'.
[0076] 2. Construction of the carrier pHZ1-CAS9-gRNA-CENH3
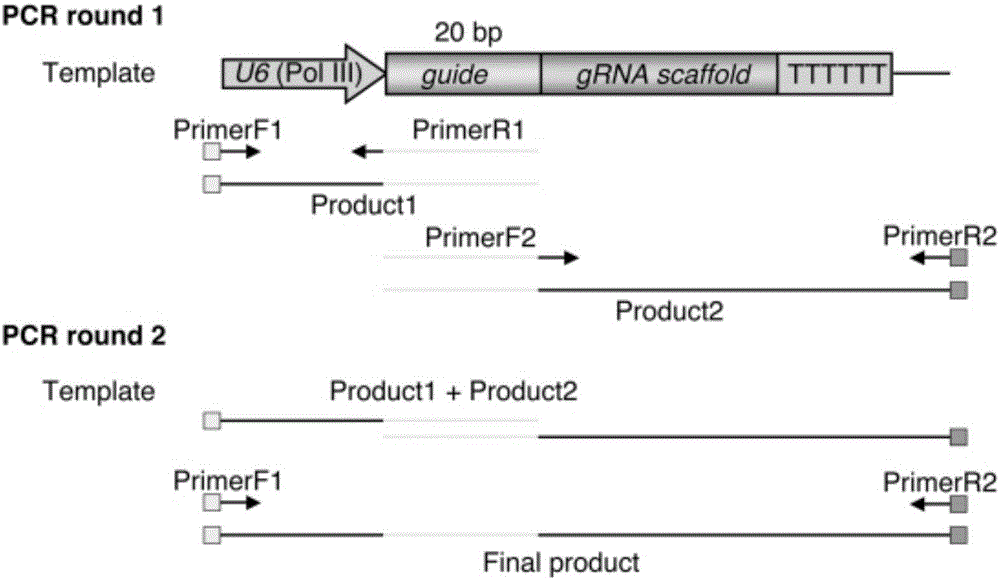
[0077] 1) Construction of OsU6-CENH3-gRNA-polyT fragment
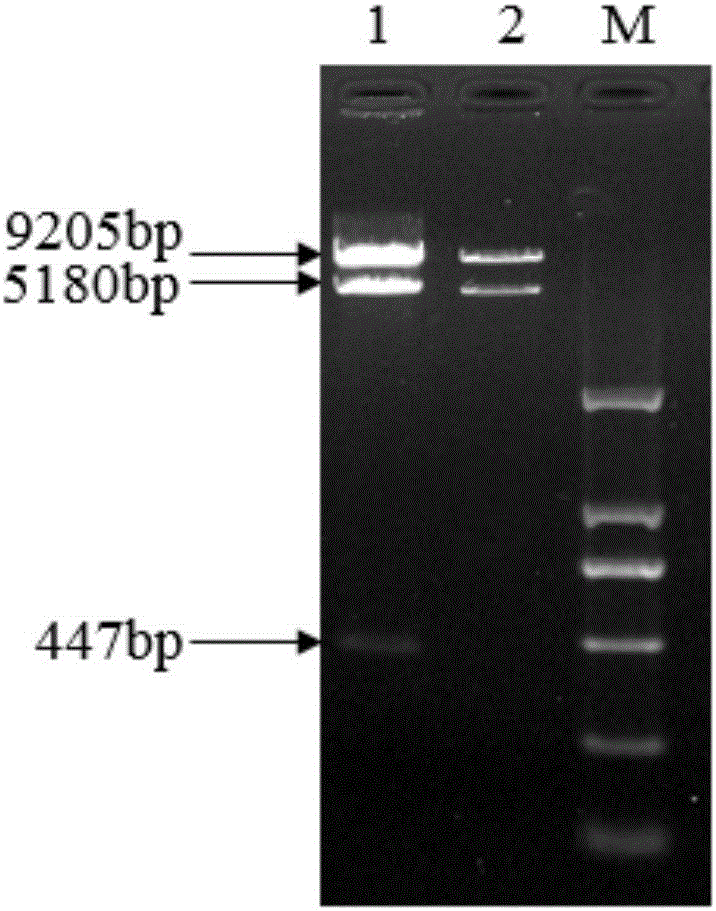
[0078] In the first round of PCR, the Cris-GL3 plasmid was used as a template, and the primers CRI-F1 and SQ2-2-R1 were used to amplify the 20bp target sequence of the OsU6 promoter and the CENH3 gene, and the fragment size was 329bp; Primers SQ2-2-F2 and CRI-R2 amplify the 20bp target sequence of CENH3 gene and gRNA-polyT, the fragment size is 180bp; the second round of PCR uses the first r...
Embodiment 2
[0088] Example 2 Preparation method of rice CENH3 gene site-directed mutation plant
[0089] 1. Agrobacterium transformation and colony PCR identification
[0090] Take the competent Agrobacterium from the -80°C refrigerator and thaw it on ice, add 3-5 μL of plasmid pHZ1-CAS9-gRNA-CENH3 and mix well, put it on ice for 30 minutes, put it in liquid nitrogen for 1 minute, and then heat shock it in a 37°C water bath 1min, add 1mL of YEP medium to the aseptic operating platform, shake on a shaker at 28°C for 2-4h, take it out, centrifuge at 8000rpm / min for 1min, pour off part of the supernatant in the aseptic operating platform, and spread the remaining medium evenly with a spreader. The suspension below was mixed and spread on the YEP plate added with rifampicin and kanamycin, sealed with parafilm and placed in a 28°C incubator for cultivation. After 2 days, pick a single colony, shake the bacteria at 28°C for 24-36 hours, and perform colony PCR to obtain positive colonies.
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com