A biomarker for detecting the prognosis of nasopharyngeal carcinoma and its application
A biomarker and nasopharyngeal carcinoma technology, applied in the field of biomedicine, can solve the problems of large differences in patient prognosis, avoid over-treatment, relieve economic pressure and medical resource pressure, and improve prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
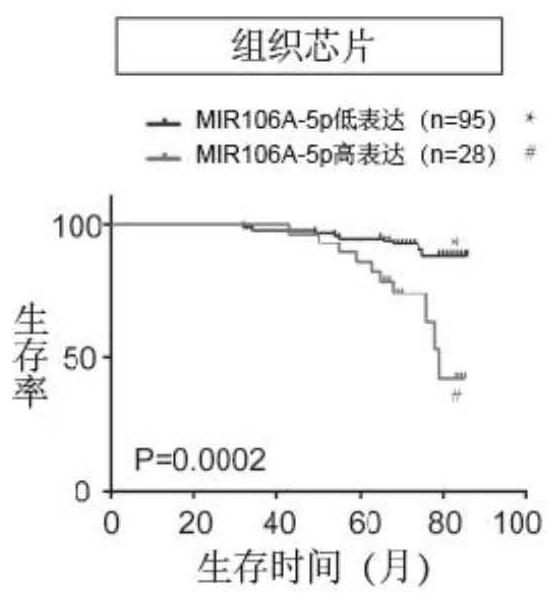
[0036] Example 1 (exploring the relationship between the biomarker miR-106a-5p and the prognosis of patients with nasopharyngeal carcinoma)
[0037] In order to explore the role of the biomarker miR-106a-5p in the progression of nasopharyngeal carcinoma, we performed an in situ hybridization (ISH) experiment on the above-mentioned 123 tissue samples, including the following steps:
[0038] Step 1: Dewax and hydrate the tissue sample chip (this step needs to be performed in a fume hood)
[0039] Soak the tissue sample chip in xylene for 30 minutes, and replace the xylene every 10 minutes; then fully soak the tissue sample chip soaked in xylene in 100% ethanol, soak in 100% ethanol for 5 minutes, and fully soak in 96% ethanol , soak in 96% ethanol for 5 minutes, fully soak in 70% ethanol, soak in 70% ethanol for 5 minutes, and soak in PBS for 5 minutes.
[0040] Step 2: Digest the tissue sample chip with protease
[0041] Submerge the dewaxed tissue sample chip in proteinase K...
Embodiment 2
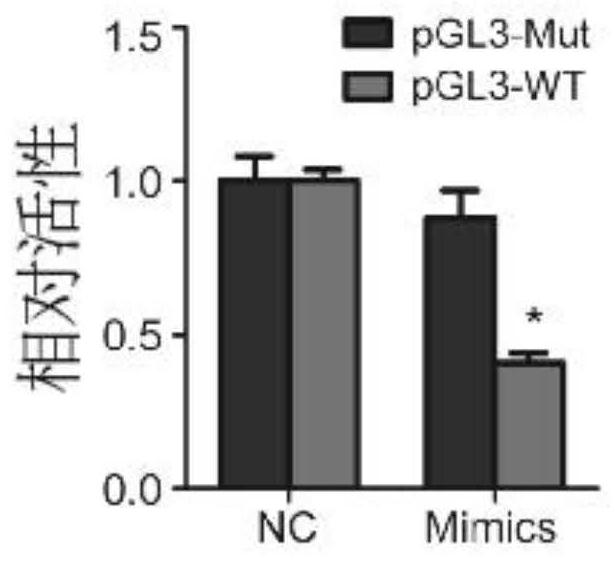
[0057] Example 2 (demonstration that the biomarker BTG3 is a direct target of miR-106a-5p)
[0058] The biologically effective interaction between miR-106a-5p and BTG3 was confirmed by the luciferase reporter gene experiment. The specific operation steps of the luciferase reporter gene experiment are as follows:
[0059] Step 1: Transfection of plasmids into nasopharyngeal carcinoma CNE2 cells
[0060] 1. Experimental group:
[0061] Group A: Plasmids are 106NC and PGL3-Mut
[0062] Group B: Plasmids are 106NC and PGL3-WT
[0063] Group C: Plasmids are 106Mimics and PGL3-Mut
[0064] Group D: Plasmids 106Mimics and PGL3-WT
[0065] 2. Inoculate CNE2 cells into a six-well plate, set 3 replicates in each group, and then inject medium into the six-well plate for cell culture. When the cell density reaches 40%-50%, add transfection reagents and corresponding plasmids for co-transfection.
[0066] Co-transfection process: Add 5 μL of Lip3000 transfection reagent to 1 mL of ba...
Embodiment 3
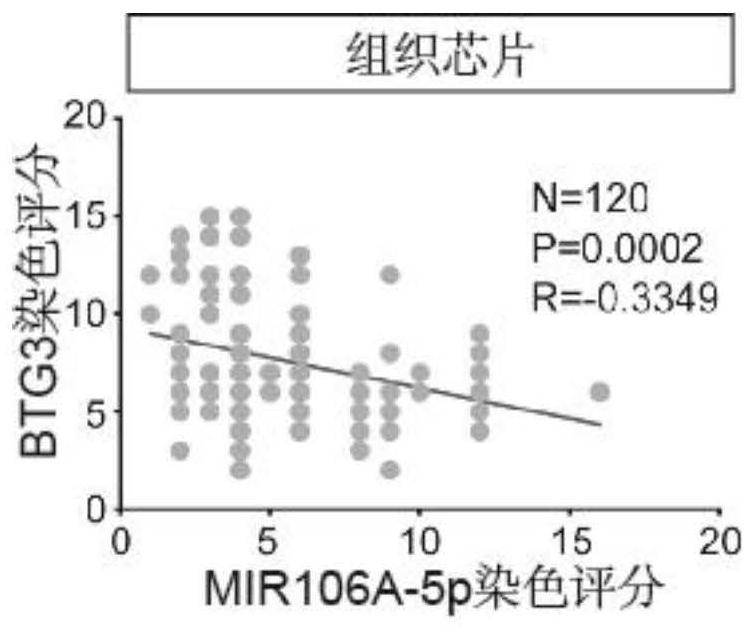
[0072] Example 3 (the relationship between the biomarker BTG3 and the prognosis of patients with nasopharyngeal carcinoma)
[0073] experimental method:
[0074] In order to explore the role of the biomarker BTG3 (i.e. miR-106a-5p target gene) in the progression of nasopharyngeal carcinoma, immunohistochemical experiments were also performed on 123 tissue samples for BTG3, including the following steps:
[0075] Step 1: Dewax and hydrate the tissue sample chip (this step needs to be performed in a fume hood)
[0076] Soak the tissue sample chip in xylene for 30 minutes, and replace the xylene every 10 minutes; then fully soak the tissue sample chip soaked in xylene in 100% ethanol, soak in 100% ethanol for 5 minutes, and fully soak in 96% ethanol , soak in 96% ethanol for 5 minutes, fully soak in 70% ethanol, soak in 70% ethanol for 5 minutes, and soak in PBS for 5 minutes.
[0077] Step 2: Antigen retrieval, quenching, blocking
[0078] 1. Heat 0.01 sodium citrate buffer s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com