SNP site related to rapid growth of large-sized takifugu rubripes fries and application of SNP site
A redfin puffer and site technology, applied in the field of fish genetic selection and breeding, can solve the problems of natural resource decline, germplasm degradation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
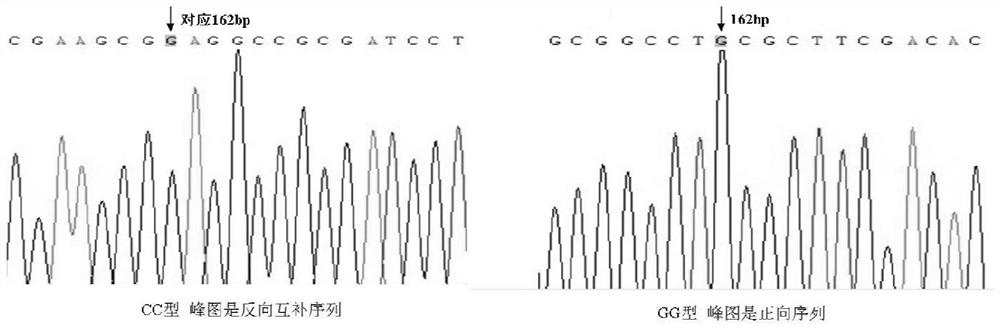
[0023] Example 1 Screening SNP sites
[0024] The screening steps of the SNP site of the present invention are as follows:
[0025] a) Extraction of the redfin fugu genome: Genomic DNA was extracted from the redfin fugu muscle by the phenol-imitation extraction method. Take 100mg of its muscle and put it into a 1.5ml centrifuge tube, and use ophthalmic scissors to cut it into pieces as much as possible, add 500ul extract and 3μl proteinase K, and mix well; after mixing, put the centrifuge tube into a 55°C water bath for water bath Heat and turn over every 15 minutes. After the sample is cracked into a clear viscous liquid, take it out and add an equal volume of saturated phenol, and mix gently for 10 minutes. Centrifuge at 12000rpm for 10min; take the supernatant, add an equal volume of phenol / chloroform (1:1) mixture and mix gently for 10 minutes, put it in a centrifuge, and centrifuge at 12000rpm for 10min; take the supernatant, add an equal volume of Chloroform was mixed ...
Embodiment 2
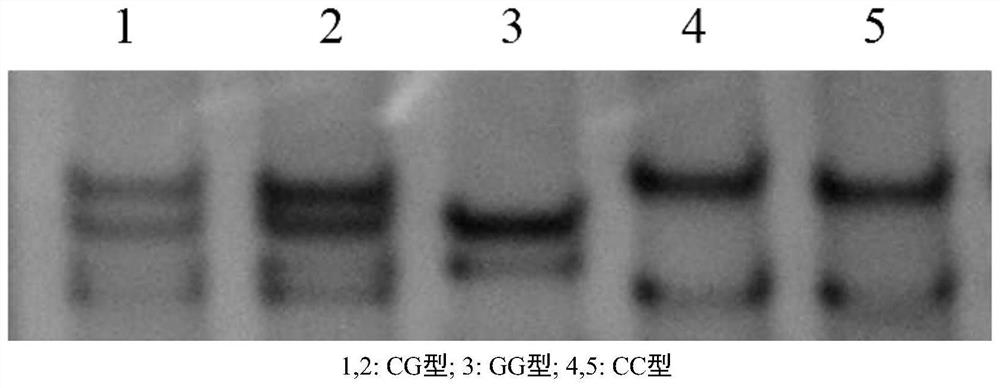
[0032] Example 2: The PCR-SSCP method proves the correlation between SNP loci and growth traits
[0033] In order to determine the SNP site associated with the growth traits of the red fin puffer fragment whose sequence is SEQ ID NO: 1, the PCR-SSCP method detection is carried out, including the following steps:
[0034] a) Acquisition of red fin puffer samples: All the experimental sample fish used were collected from Dalian Fugu Aquaculture Co., Ltd., and were grown into randomly selected 102-day-old red fin puffer under the same breeding management and nutritional conditions There are 296 individual fry.
[0035] b) Data collection and genomic DNA extraction: the phenotype values of body weight and body length of 296 individuals were measured and recorded, and muscle tissue was collected for genomic DNA extraction.
[0036] c) PCR reaction and sequencing. The reaction system is 25 μl: 10×buffer 2.5 μl, dNTP 2 μl, F primer (SEQ ID NO: 2) 1 μl, R primer (SEQ ID NO: 3) 1 μl,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com