Molecular marker SNP#2 for identifying powdery mildew resisting phenotype of vigna radiata, and primer and application of molecular marker SNP#2.
A molecular marker and anti-powdery mildew technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problem that molecular markers closely related to resistance have not been reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
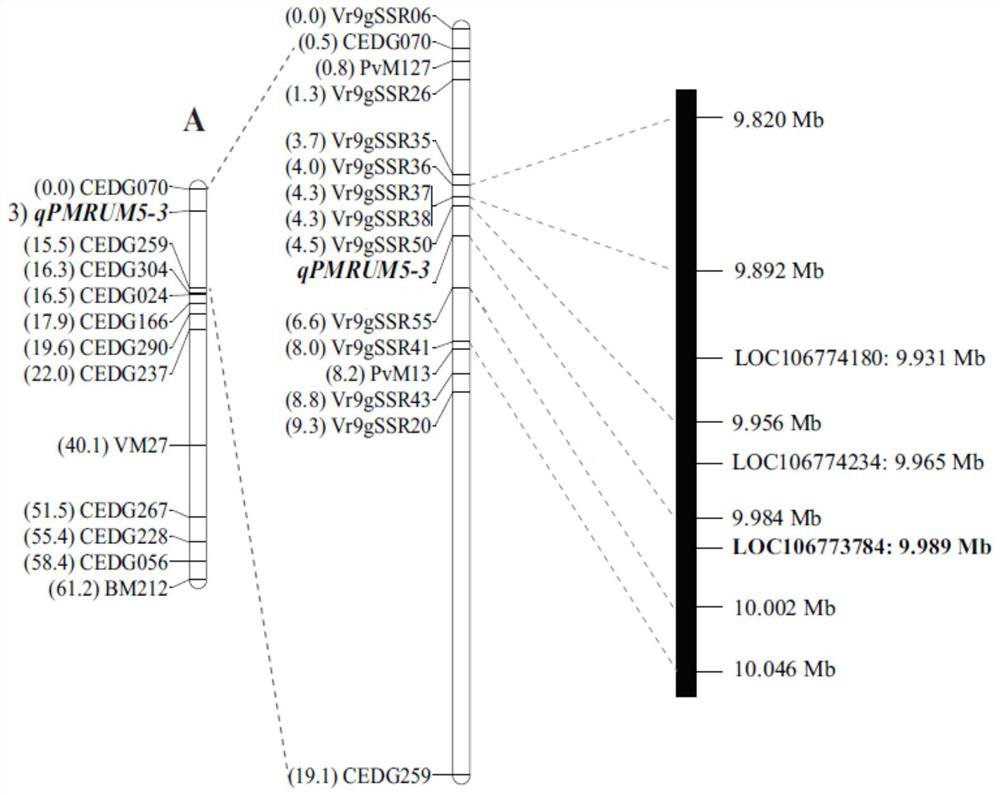
[0025] Example 1 CN60 X RUM5 F2 population 190 strains, CN60XRUM5 BC1F1 population 74 strains, population construction and phenotypic identification
[0026] Using the wild powdery mildew-resistant mung bean resource RUM5 as the male parent and the powdery mildew-susceptible mung bean resource CN60 as the female parent, hybrids were prepared, and an F2 segregation population containing 190 individuals was constructed. Each F2 individual plant obtained the corresponding F2 by selfing : 3 families were identified for resistance to powdery mildew. In addition, the wild powdery mildew-resistant mung bean resource RUM5 was used as the male parent, and the powdery mildew-susceptible mung bean resource CN60 was used as the female parent to prepare hybrids, and a BC1F1 segregation population containing 74 individuals was constructed, and each individual plant was self-crossed to obtain a BC1F1: 2 family , carry out powdery mildew resistance identification, two groups of synchronous te...
Embodiment 2
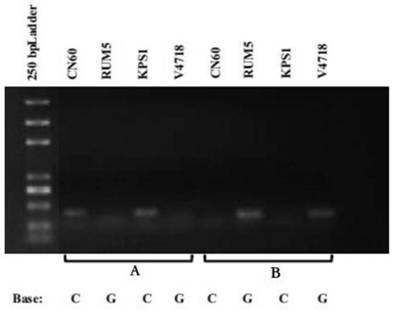
[0037] Extract the mung bean powdery mildew susceptible materials KPS1, Sulu 1, Sulu 3, powdery mildew resistant materials V4718, Sulu 2, V2802 and V2709, and the foreign mung bean wild species TC1966 genomic DNA, and use this molecule to mark the primer of SNP#2 (SEQ ID NO.1 / SEQ ID NO.2 and SEQ ID NO.3) were amplified by PCR for powdery mildew resistance identification, the results are shown in Table 2, and the results showed that the identification efficiency of the marker was 100%.
[0038] Table 2
[0039]
[0040] The identification of mung bean powdery mildew resistance by the above molecular markers can predict the disease resistance of mung bean plants, which can improve the breeding process of mung bean powdery mildew resistant varieties in my country.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com