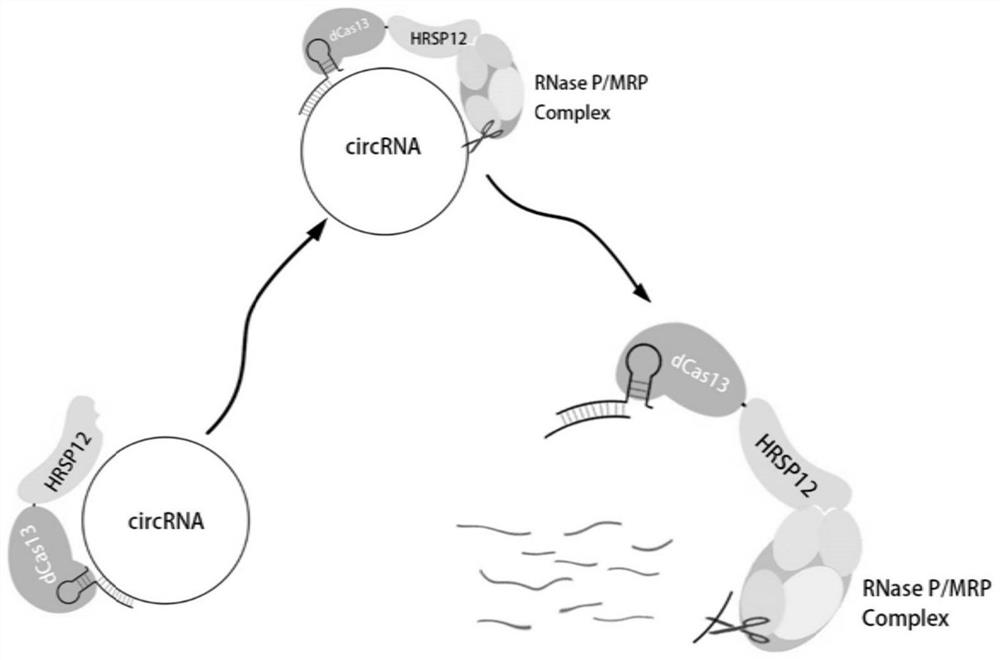
Circular RNA knockdown method and application thereof
A circular and protein technology, applied in the field of molecular biology, can solve the problems of non-specific targeting of parental RNA, low efficiency and off-target when circular RNA is knocked down, and achieve high-efficiency specific targeting, high design selectivity, and off-target less effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1 dCas13-HRSP12 vector construction
[0053] 1. PCR amplification of the target fragment
[0054] Using LwaCas13a-91902 plasmid, 293T cell cDNA and PCDH-3xFlag plasmid as templates, respectively amplified inactivated dLwaCas13a partial sequence D1 to 1459bp, inactive dLwaCas13a partial sequence D2 to 1735bp, inactive dLwaCas13a partial sequence and Linker and NES sequence D3 is 418bp, Linker and HRSP12 sequence D4 is 430bp, protein tag 3xFlag sequence D5 is 147bp, the specific sequence is as follows:
[0055] KD131-D1-UnF:GCTGGCTAGCGTTTAAACTTAAGCTTGCCACCATGAAAGTGACCAAGGTCGA
[0056] KD131-mut1-R:CCGTGAGCAATACTAGAGATGGCCTCATCGATATTGGCGAAGAAGTCCTCGATCT
[0057] Amplified size 1459bp;
[0058] KD131-mut1-F:CCATCTCTAGTATTGCTCACGGCATCGTGCATTTCAACCTGGAACTGGAAGGCAAG
[0059] KD131-mut2-R:GTTTGCAATATACAGGTCTTTTCTTCTCCTGCTTCAGTTTCTTCACTTTCT
[0060] The amplification size is 1735bp;
[0061] KD131-mut2-F:GAAGAAAGACCTGTATATTGCAAACTACATCGCTCACTTTAATTATATTCCTCAT
...
Embodiment 2
[0105] Embodiment 2 The design of gRNA and the construction of vector
[0106] Taking dLwaCas13a (KDHP-a) as an example, the design of the supporting gRNA and the construction of the vector include the following steps:
[0107] 1. Design of gRNA supporting dLwaCas13a
[0108] The gRNA supporting dLwaCas13a consists of DR plus target sequence, in the form of GATTTAGACTACCCCAAAAACGAAGGGGACTAAAACNNNNNNNNNNNNNNNNNNNNNNNNN, where N is the gRNA sequence.
[0109] The primer design software was used to retrieve the sequences at the circularization sites of circular RNAs circPVT1 and circHIPK3, and parameters such as GC content were calculated. The gRNA target sequences were designed as follows, and the corresponding vectors were named G11-G16 and G21-G27, respectively.
[0110] >gRNA targrt-circPVT1-1(11+11)
[0111] CTGGGCTTGAGGCCTGATCTTT
[0112] Among them, the brackets indicate the number of upstream bases at the cyclization site plus the number of downstream bases, that is, 1...
Embodiment 3
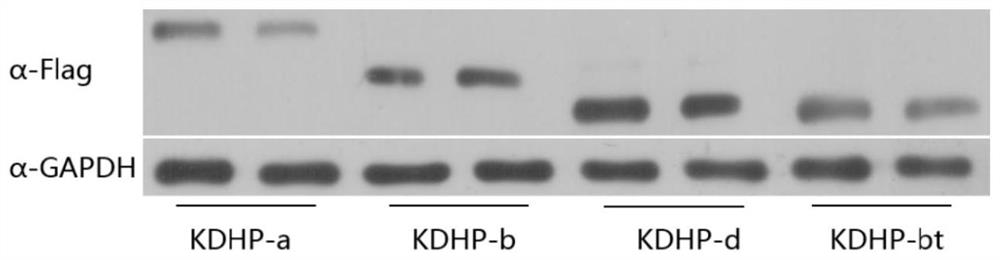
[0186] Example 3 Vector transfection and expression of KDHP protein
[0187] Using Lipofectamine 3000, dLwaCas13a (KDHP-a), dPspCas13b (KDHP-b), dRfxCas13d (KDHP-d) and dPspCas13b-truncated (983aa) (KDHP-bt) vectors were used to transfect 293T cells, and the samples were digested and centrifuged after 48 hours.
[0188] Wash 3 times with pre-cooled PBS, blot the residual liquid as much as possible, add 100 μL of cell lysate containing PMSF, lyse at low temperature for 20 minutes, and transfer the liquid to a 1.5 mL EP tube. Then add 5×Loading buffer (at a ratio of 2:1), boil at 100°C for 10 minutes and perform SDS-PAGE electrophoresis.
[0189] Using PVDF membrane, transfer membrane at 350mA for 80min. Then soak the membrane in blocking solution at 37°C for 1 h; take out the membrane and put it directly into the primary antibody (Flag antibody diluted 3000 times), overnight at 4°C; wash the membrane with PBST, 5min×4 times; transfer the membrane to the secondary antibody ( D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com