Downregulation of snca expression by targeted editing of dna-methylation
A targeted, gene-based technology for DNA/RNA fragmentation, recombinant DNA technology, stable introduction of foreign DNA into chromosomes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0182] Materials and methods
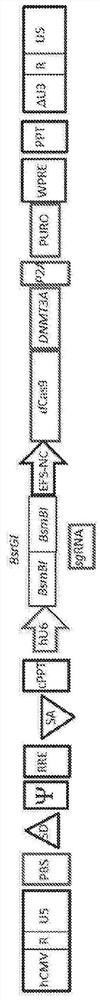
[0183] Plasmid design and construction. The dCas9-DNMT3A transfer gene was derived from pdCas9-DNMT3A-EGFP (Addgene plasmid number 71666), and cloned into pBK301 (production-optimized lentiviral vector) as follows: by cloning the dCas9 fragment digested with AgeI-BamHI restriction enzymes Into pBK301, the pBK456 plasmid was generated. Next, the DNMT3A catalytic domain was transferred from pdCas9-DNMT3A-EGFP into pBK456 by amplifying a DNMT3A fragment from a plasmid using primers containing a BamHI restriction site: BamHI-429 / R 5'-GAGCGGATCCCCCTCCCG-3' (SEQ ID NO: 15), BamHI-429 / L 5'-CTCTCCACTGCCGGATCCGG-3' (SEQ ID NO: 16). The pBK456 was then digested with BamHI restriction enzyme for cloning, resulting in pBK492 plasmid (no gRNA plasmid). Next, the extra BsmBI site located in the DNMT3A fragment was eliminated by site-directed mutagenesis to generate pBK546 (SEQ ID NO:39; see Figure 12B ). This plasmid contains dCas9-DNMT3A-p2a-puromycin exp...
Embodiment 2
[0214] Development of a novel lentiviral vector system for efficient delivery of CRISPR / Cas9-based epigenetic tools
[0215] One disadvantage of the all-in-one integrative lentiviral vector system for the delivery of CRISPR / Cas9-based materials is low production titers. Approaches to overcome these issues include the development of binary plasmid vector systems that deliver the Cas9 and gRNA components separately. This approach has improved yield but is not suitable for gene editing applications including in vivo screening and disease modeling. Recently developed second-generation all-in-one vectors have shown increased production titers and transduction efficiencies compared to first-generation systems, but they still yield ~25-fold lower yields compared to traditional vectors. The ability to simultaneously deliver Cas9 and sgRNA via a single vector enables easy and robust gene editing in vivo, which is particularly advantageous for the development of translatable gene thera...
Embodiment 3
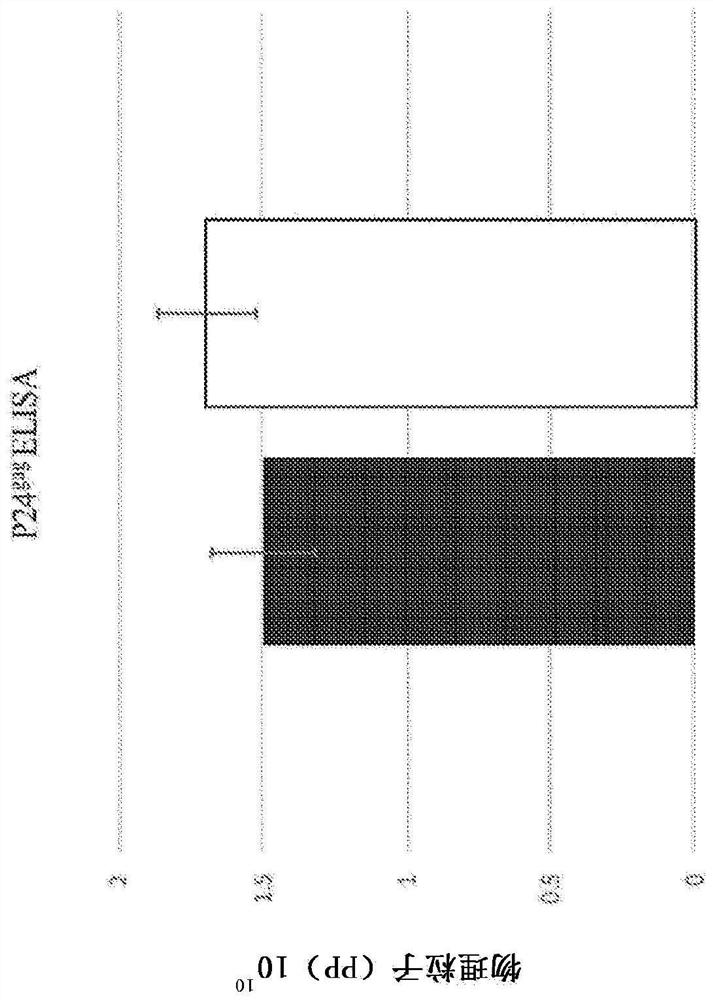
[0218] Results - Targeted methylation of SNCA-intron 1 using the all-in-one lentiviral vector-dCas9-DNMT3A system
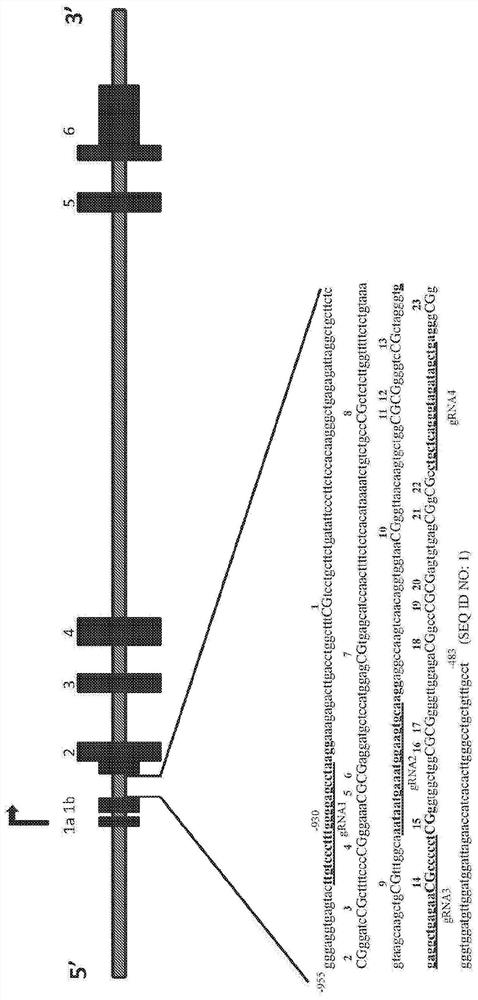
[0219] SNCA intron 1 contains a CpG island (CGI) region containing 23 CpGs [Chr4: 89,836,150-89,836,593 (GRCh38 / hg38)] ( Figure 1A), where the methylation status changes with increasing SNCA expression. In addition, SNCA intron 1 subregions may be differentially methylated in disease states. CpG sites in this subregion of intron 1 may serve as candidate targets for epigenetic manipulations related to the fine regulation of SNCA transcription, and enhanced DNA methylation in these CpG sites may allow the regulation of SNCA expression. Stringent downregulation and reversal of PD-associated phenotypes. To assess this hypothesis, an all-in-one gRNA-dCas9-DNMT3A lentiviral vector was constructed using a production and expression-optimized backbone containing repeats of the transcription factor Sp1 binding site upstream of the human U6 (hU6) promoter and a present ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com