Method for predicting protein and ligand molecule binding free energy based on convolutional neural network
A technology of convolutional neural network and ligand molecules, which is applied in the field of predicting the binding free energy of proteins and ligand molecules based on convolutional neural networks, can solve problems such as limited application, difficult generalization of homologue ligands, and difficulties in large-scale application , to achieve the effects of small error, good generalization ability, and precise protein-ligand binding free energy calculation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Binding free energy prediction for a kinase target:
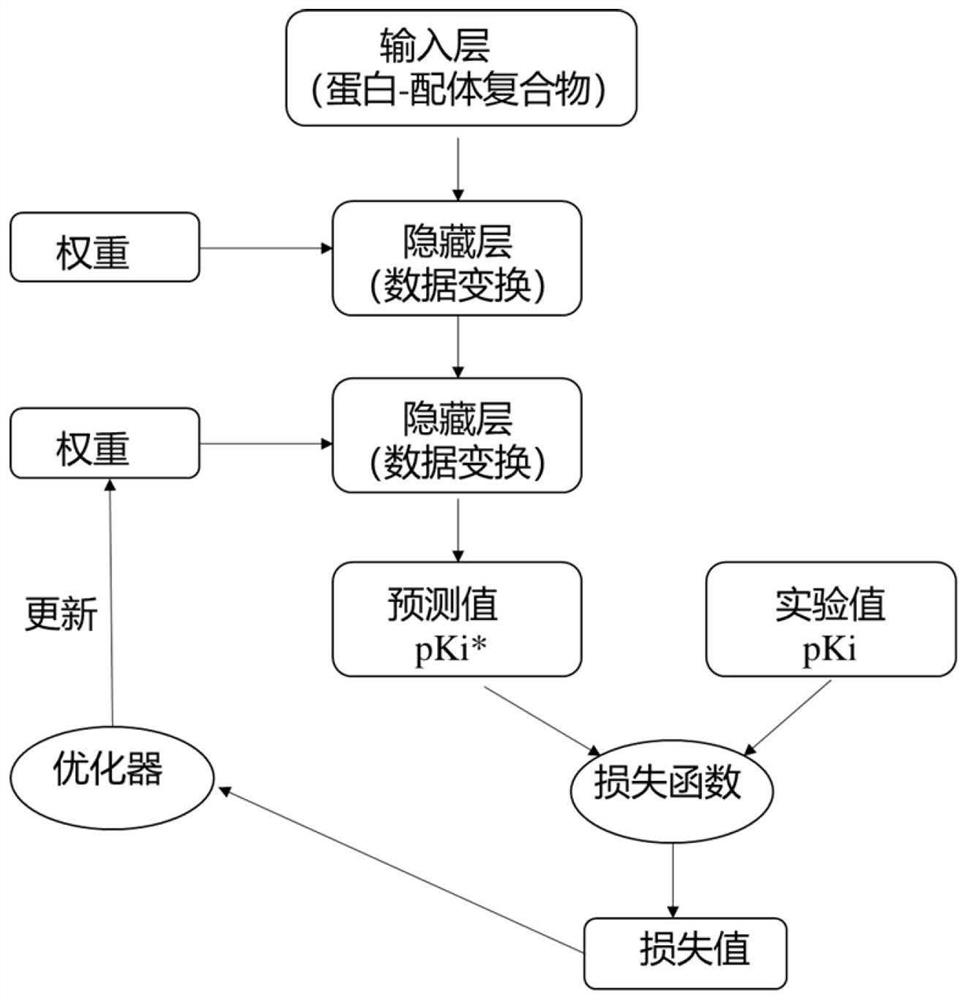
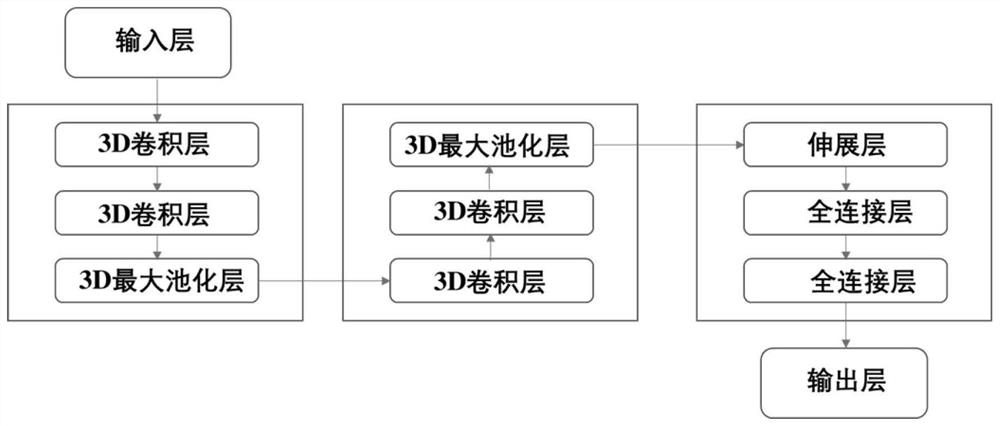
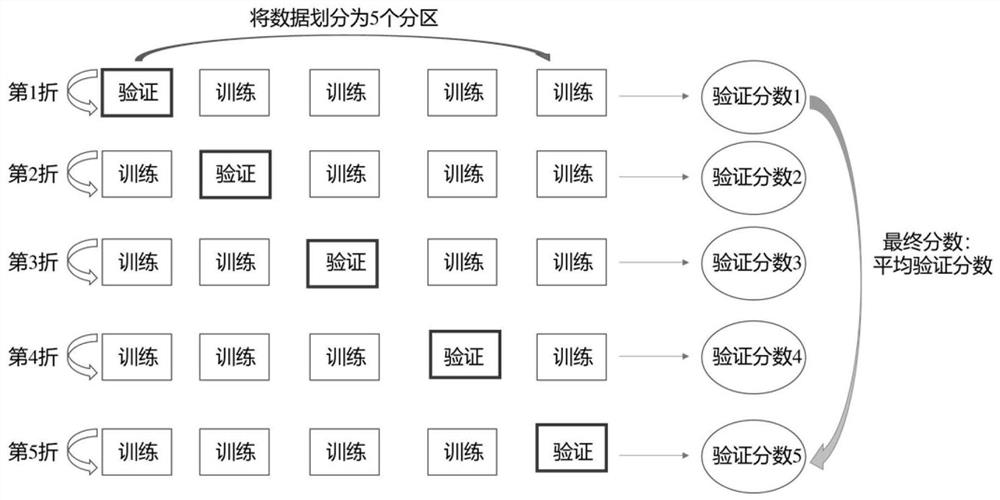
[0050] First, collect the small molecule inhibitors of the target, perform 3D conformation preparation and molecular docking calculation, and then call the molecular descriptor module to process these molecules and proteins and input them into the convolutional neural network model. Such as image 3 As shown, all the molecular data are divided into 5 parts, one of which is selected as the verification set each time, and the other four are used as the training set, so that the model will be trained for 5 rounds in total, that is, 5-fold cross-validation, and finally all the verification scores will be taken as The average is used as the final validation score. Figure 4 , Figure 5 Indicates the performance on the verification set during model training, from Figure 4 It can be seen from the figure that the error in the initial stage of the model is relatively large, but after about 50 rounds of training, the error...
Embodiment 2
[0054] Lead compound structure optimization for a target:
[0055] Some seed compounds with good initial activity were obtained through virtual screening, and the convolutional neural network model was used to optimize the structure of this batch of seed compounds to obtain lead compounds.
[0056] Molecular docking of hit compounds and targets is first performed, and then they are encoded and molecular descriptors are calculated. Input it into the convolutional neural network model to predict the binding free energy of the compound to the target. According to the binding mode between the hit compound and the target, as well as the binding free energy value and spatial hierarchy information predicted by the model, the structure of the hit compound is modified and optimized, and groups matching the spatial hierarchy of the target are added to the hit compound , form a better structural complementarity with the target to design new compounds. Afterwards, molecular docking and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com