Primers and method for high-efficiency detection of ACDs genome-wide loci based on NGS sequencing
A genome sequence and insertion site technology, applied in the field of molecular biology, can solve the problems of low efficiency, restricted development, high cost, etc., and achieve the effect of reducing cost and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1A
[0031] Embodiment 1Ac / Ds specific primer design and inspection
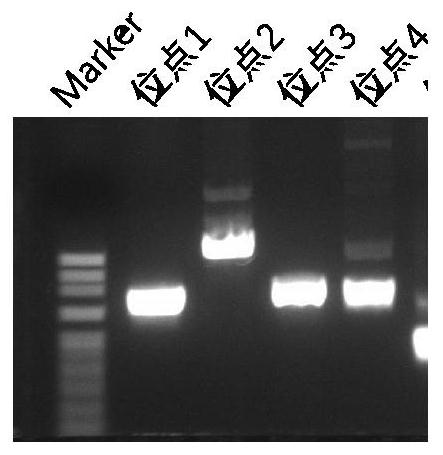
[0032] In the present invention, according to the Ds sequence whose genbank seqID in NCBI is JF421358.1 as a reference, scanning and alignment are performed in the genome of maize B73 to obtain 73 Ac / Ds. According to the consensus sequence characteristics of these Ac / Ds, each 150bp conserved sequence at the end was cut out, and the comparison results are shown in Figure 5 . In the present invention, all Ac / Ds sites at the genome-wide level will be obtained finally by NGS sequencing technology. At present, the read length of NGS is generally 150 bp at the single end, and the sum of the double ends is 150-300 bp, which is a short read length. In order to ensure that the transposon sequence greater than 50bp and the adjacent genome sequence greater than 50bp can be obtained within this read length range, the present invention designs the 5' or 3' end primers on the Ac / Ds to be about 70bp away from the ends of the...
Embodiment 2
[0037] Example 2 Enrichment of Ac / Ds loci from the maize genome by a high-throughput NGS sequencing method
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com