Method for predicting glioma molecular subtypes and prognosis based on depth image features
A molecular subtype and depth image technology, applied in image analysis, image enhancement, image data processing, etc., can solve the problems of expensive, invasive, etc., and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] A method for predicting the molecular subtype and prognosis of glioma based on deep image features, comprising the following steps:
[0052] Step 1, constructing a data set: the data set includes magnetic resonance imaging (MRI) of multiple cases of primary glioma patients.
[0053] Step 2, build a feature extractor based on RA-UNet neural network:
[0054] 2-1. Use RA-UNet to segment the magnetic resonance imaging MRI of each patient with primary glioma in step 1 to generate segmentation results, and generate n pieces of magnetic resonance imaging MRI for each patient with primary glioma Split results.
[0055] 2-2. For the segmentation results obtained in step 2-1, use the feature extractor to extract high-dimensional features. The high-dimensional feature dimensions extracted from each segmentation result are (64, 64, 64, 32), integrated into each case The high-dimensional feature dimensions of MRI in primary glioma patients are (n, 64, 64, 64, 32);
[0056] Step ...
Embodiment 2
[0070] A method for predicting the molecular subtype and prognosis of glioma based on deep image features, comprising the following steps:
[0071] Step 1, data preprocessing
[0072] 1-1 Prepare the data set, which comes from the pretreatment MRI of primary glioma patients in The Cancer Genome Atlas. Each MRI includes four modes of T1, T1Gd, T2, and FLAIR, that is, four scan modes .
[0073] Molecular subtype information and survival data for glioma patients were obtained from cBioPortal, a public domain clinical and molecular data repository of The Cancer Genome Atlas patient cohorts.
[0074] In the embodiment of the present invention, the data set has a total of 71 cases, that is, the pretreatment MRIs of 71 patients with primary glioma. Among them, the four molecular subtypes of classical, neuronal, proneural and mesenchymal included 15, 9, 18 and 29 patients, respectively. We divided patient survival into short (short) - less than 6 months, medium (medium) - 6 to 18 m...
Embodiment 3
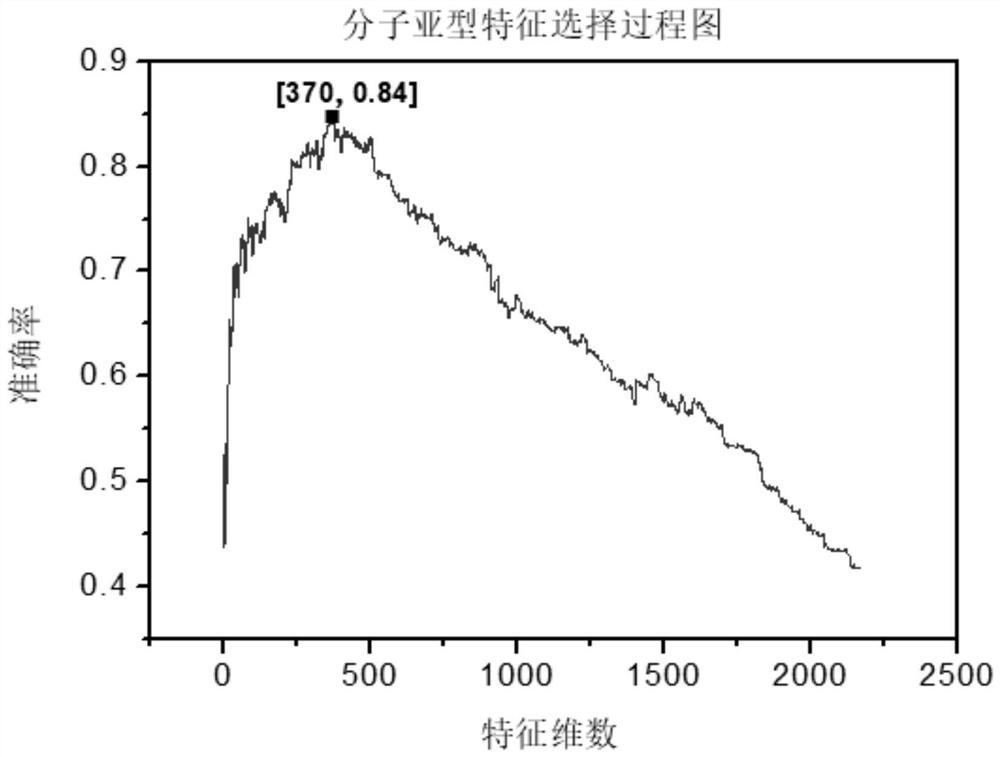
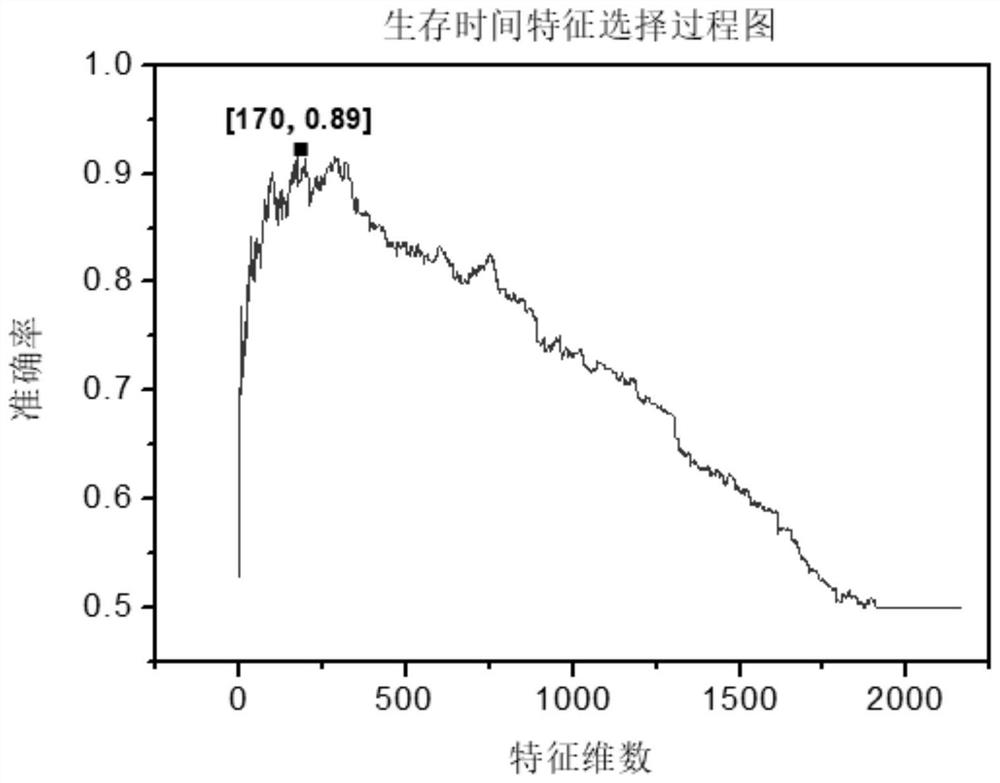
[0099] We sort the 2166 features obtained in step 3-3 in descending order according to their F-score values, and set the selected feature set as an empty set initially, and select a feature with the largest F-score value from the unselected features to add To the selected feature set, and then use the selected feature set to train SVM, each training will get an accuracy rate, until 2166 features are added to the selected set, and the iteration ends. Finally, experimental data show that for molecular subtype prediction problems such as figure 2 As shown, the number of features achieves the best effect when the number of features is 370, and the accuracy reaches 82%. In the survival time prediction problem, such as image 3 As shown, the number of features achieves the best effect when the number of features is 174, and the accuracy reaches 94.8%.
[0100] The training process of the SVM classifier adopts the method of 10-fold cross-validation. For simplicity and intuition, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com