High-throughput sequencing library construction method of nucleic acid aptamer library
A nucleic acid aptamer, sequencing library technology, applied in chemical libraries, DNA preparation, combinatorial chemistry, etc., can solve the problems of PCR amplification preference, primer residues, etc., to avoid base preference, increase output and analysis, To achieve the effect of complete retention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 High-throughput sequencing library construction method of nucleic acid aptamer library (1)
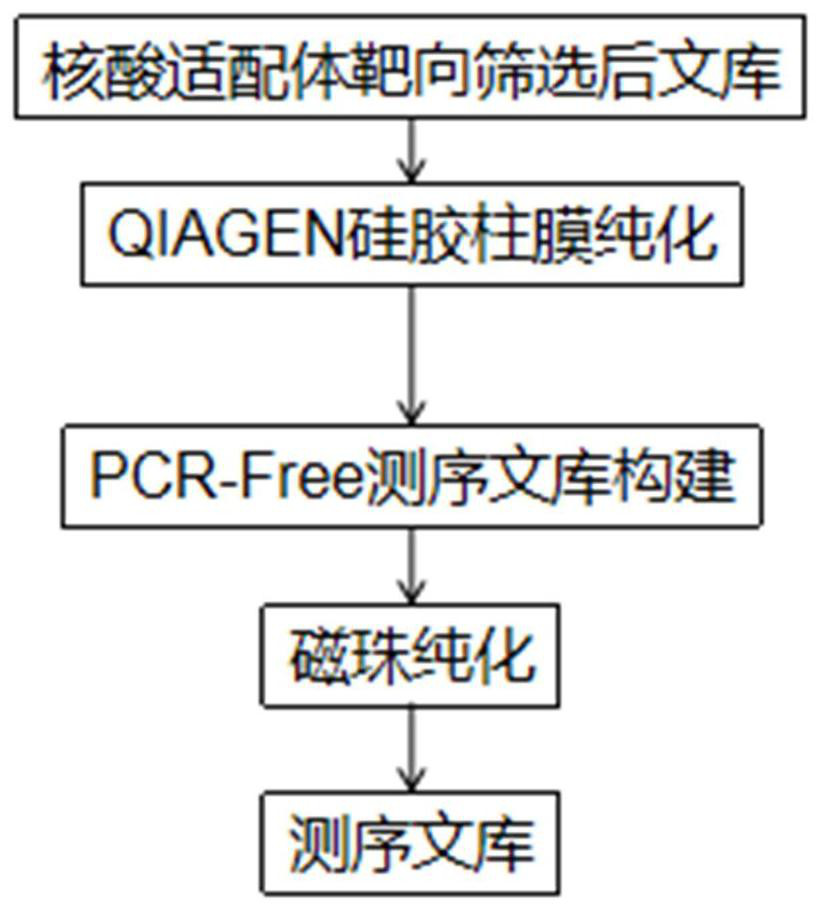
[0048] This embodiment provides a method for constructing a high-throughput sequencing library of a nucleic acid aptamer library. The schematic flow chart of the method is as follows: figure 1 As shown, it specifically includes the following steps:
[0049] 1. Use the QIAGEN MinElute PCR Purification Kit to purify the nucleic acid aptamer PCR product mixture library
[0050] (1) Take 5 times the volume of PB of the sample to be purified and mix it with the sample to be purified, add it to the silica gel membrane purification column, place the silica gel membrane purification column on a 2mL collection tube, and let it stand for 2 minutes, so that the nucleic acid and the column membrane are fully combined.
[0051] (2) Centrifuge at 8000rpm for 1min, and discard the waste liquid.
[0052] (3) Add 750 μL PE (absolute ethanol has been added and mix well) to the purifi...
Embodiment 2
[0066] Example 2 High-throughput sequencing library construction method of nucleic acid aptamer library (2)
[0067] This example provides a method for constructing a high-throughput sequencing library of a nucleic acid aptamer library, which differs from the method in Example 1 only in that in step 3, the reaction is performed at 24°C for 10 minutes and at 22°C for 20 minutes.
experiment example 2
[0072] Analysis of the quality output and sequencing data output of the sequencing library in Experimental Example 2
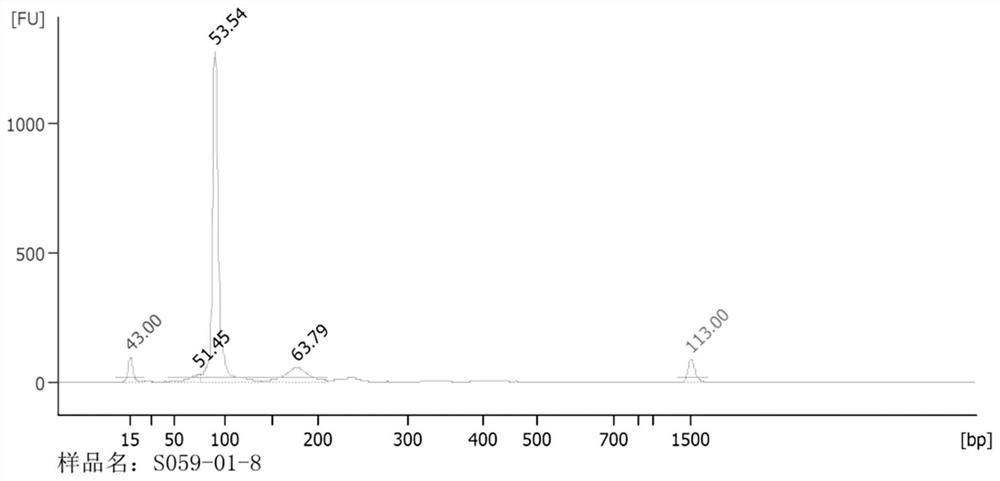
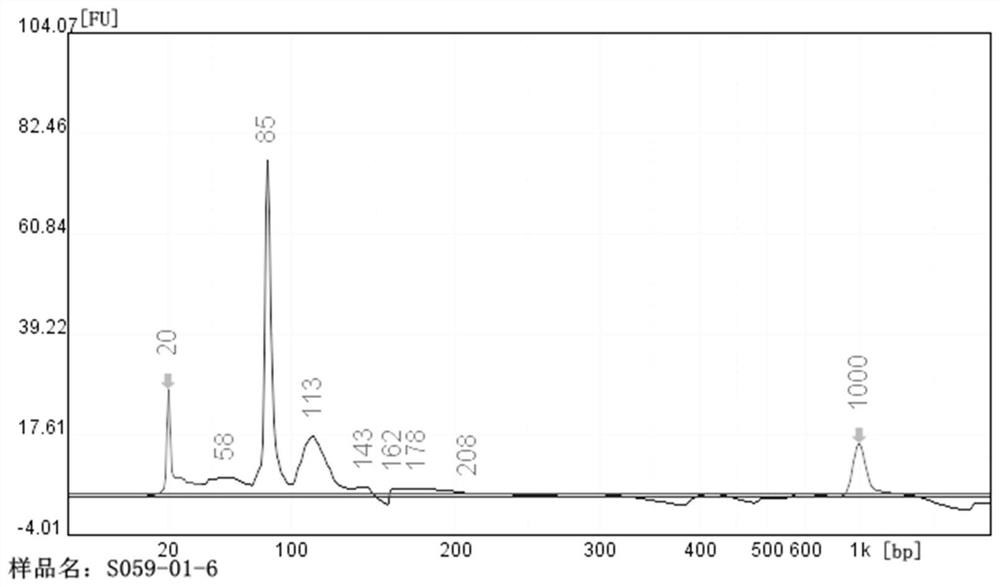
[0073] The quality output of the library and the output analysis of the sequencing data were carried out on the high-throughput sequencing library constructed by the library construction method of the above Example 1, Example 2 and Comparative Example 1. The results are shown in Table 1 and Table 2. The results show that , the sequencing library of the nucleic acid aptamer library sample constructed by the method of Example 1 has a higher concentration and about 90% of effective data, and the sequencing library of the nucleic acid aptamer library sample constructed by the method of Example 2 is higher than that of the implementation The concentration and effective data of the library in Example 1 are slightly inferior, and the effective data of sequencing can be about 80%. The effective concentration, QPCR concentration and data output of the library S059-R01 c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com