Recombinant escherichia coli constructed by genetic engineering and method for biosynthesizing 6'-sialyllactose
A technology for recombining Escherichia coli and genetically engineered bacteria, which is applied in the field of industrial microbial metabolic engineering, and can solve problems such as differences in relative reactivity and expensive sugar donors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Gene acquisition
[0073] In this embodiment, the gene of different sources is optimized, including: N-acetyl glucamine isomerase gene derived from E. coli Neuc (Optimized SEQ ID NO.1), acetyl neopropase synthase gene NEUB (Optimized SEQ ID No.2), CMP-Acetyl Neurine Synthetase Gene Neua (Optimize the gene SEQ ID NO.3), from Photobacterium Leiognathi of 2,6st Gene (optimized gene SEQ ID NO.4), derived from the phosphate amine acyllet transferase gene of brewing yeast GNA1 (Optimize the gene SEQ ID No.5), from the compositivity SYNECHOCYSTISTISTISTISTISTISTIS SP. N-acetyl glucosamine differential contrast enzyme gene AGE (Optimize the gene SEQ ID NO. 6).
Embodiment 2
[0075] Preparation of recombinant plasmid
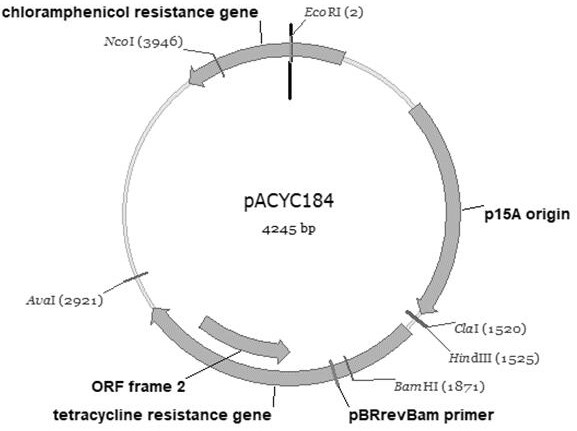
[0076] CMP-acetyl neurine synthetase gene derived from E. coli obtained in Example 1 Neua , Acetyl neurine synthetase gene NEUB , N-acetyl glucosamine isomerase gene Neuc PCR amplification was performed, and the amplified fragment was purified and purified, and the double enzyme digestion was carried out, and the fragment of the enzyme digestion was connected to the plasmid of the plasmid Pacyc184 which was also subjected to bisase. 3 proportion of mixing, adding T4 DNA Ligase to 22 ° C for 5 h at 22 ° C, connection product conversion E. Coli DH5α, screening on chloramphenicol plate, gaining recombinant plasmid PACYC- Neucba .
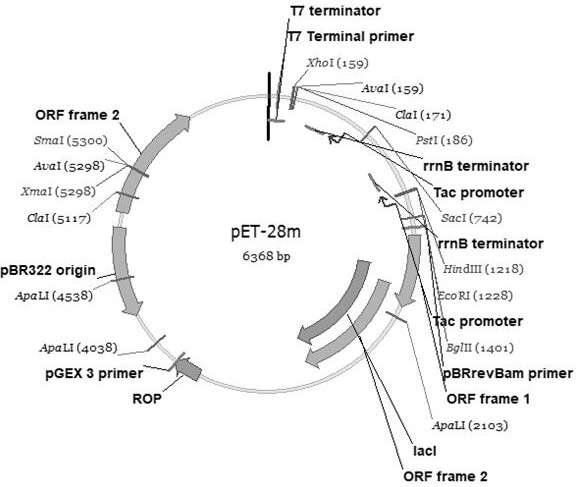
[0077] The origin obtained in Example 1 Photobacterium Leiognathi of 2,6st The gene was amplified in PCR, and the amplified fragment was purified, and the fragment of the bisase digestion was also passed through a double-cut PCYC- Neucba The plasmid is connected, and the carrier: The segment of the target...
Embodiment 3
[0081] Gene's knockout
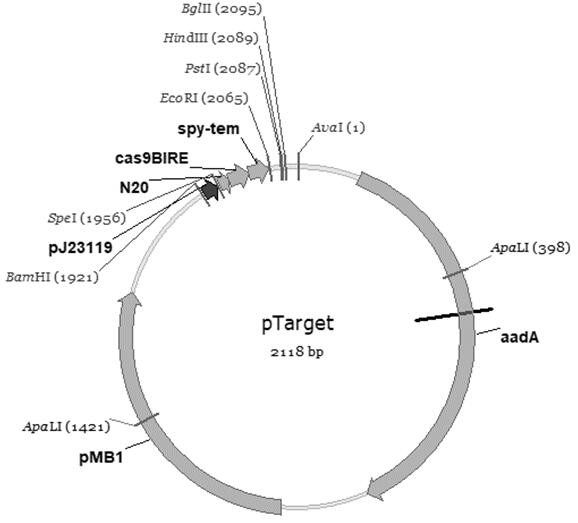
[0082] This embodiment uses RED homologous recombination assisted CRISPR-CAS9 methods to further simplify gene editing methods, enabling efficient and rapid and continuous editing of Escherichia coli genotype, rapidly obtaining design induction genetic and physiological metabolic characteristics. Below LDH Gene is an example, and the step of the gene knockout is explained in detail, and the knockout of the remaining gene is the same.
[0083] Find in NCBI E. Coli LDH Nucleotide sequence of genes, design LDH Deletion primers and identification of genes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com