Method for analyzing Granger causality among multiple florae based on pathogenic microorganism metagenome
A technology of pathogenic microorganisms and metagenomics, applied in the field of Granger causality analysis among various bacterial groups, can solve many and complex problems, limited to empirical anti-infection, pathogens cannot be clearly defined, and achieve high reliability results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0020] The following will clearly and completely describe the technical solutions in the embodiments of the present invention with reference to the accompanying drawings in the embodiments of the present invention. Obviously, the described embodiments are only some, not all, embodiments of the present invention. Based on the embodiments of the present invention, all other embodiments obtained by persons of ordinary skill in the art without creative efforts fall within the protection scope of the present invention.
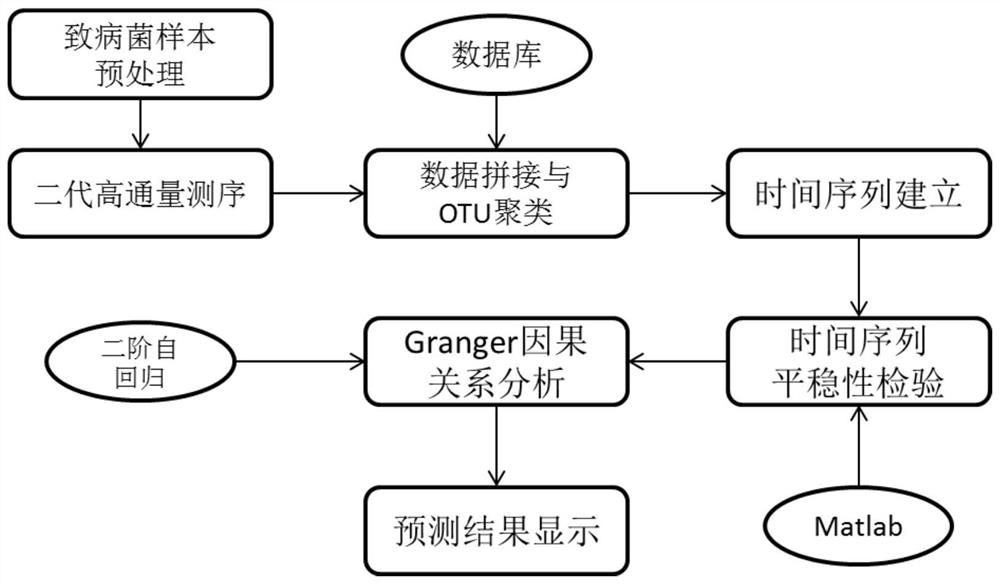
[0021] Such as figure 1 As shown, the present invention clusters and analyzes the obtained pathogenic microorganism metagenomic data and divides each strain, calculates the species abundance of each strain at the corresponding time node of disease development, and then establishes each time point for each time point. The time series of group variables is corrected, and finally Granger causality analysis is performed on the corrected data to find the main pathogenic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com