Compound target protein binding prediction method based on multi-deep learning model consensus
A learning model and deep learning technology, applied in the field of pharmaceutical research and development, can solve problems such as numerous parameters to be adjusted, high false positives, and low success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
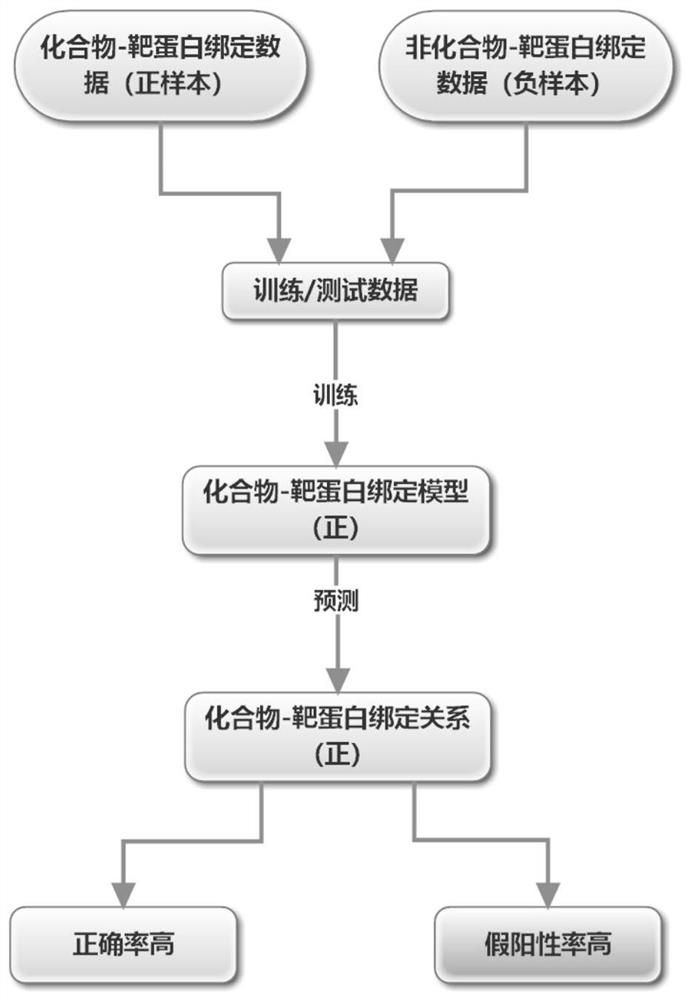
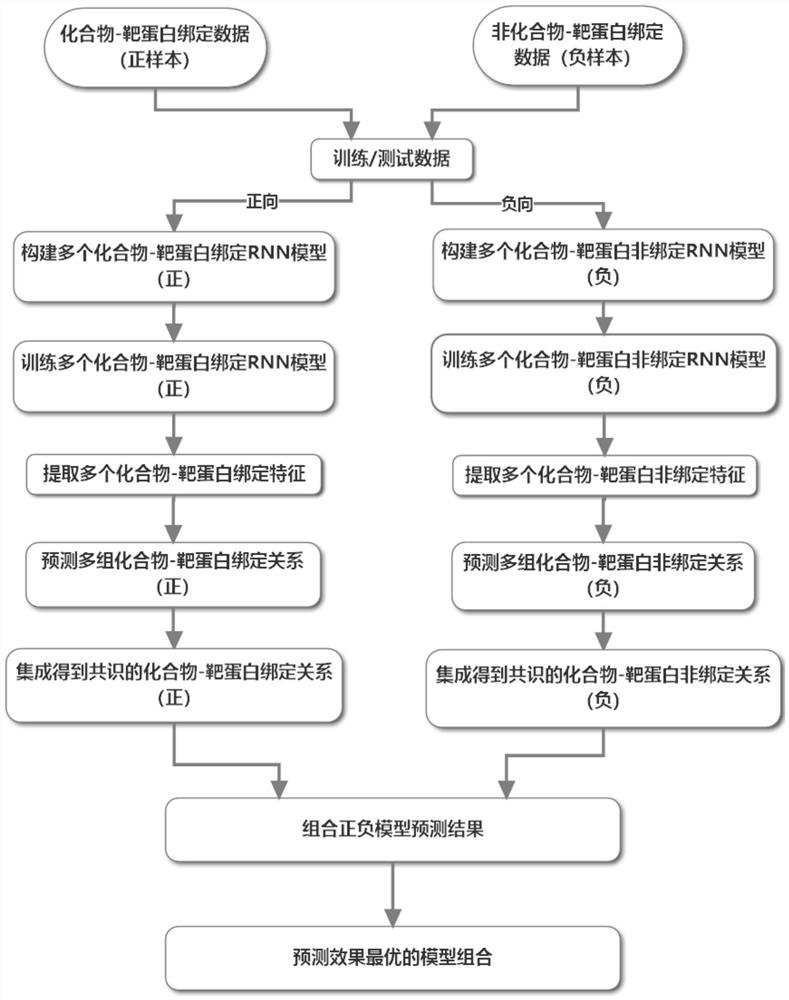
[0072] Multiple "recurrent neural network (positive)" models are connected in series to multiple "recurrent neural network (negative)" models to predict the binding relationship and system of compound target proteins.
[0073] Schematic diagram of the overall system structure: see Figure 3 ~ Figure 4 .
[0074] System operating environment:
[0075] Hardware: CPU+GPU;
[0076] Software: Windows or Linux, Python+Tensorflow, PyTorch, Keras, etc.
[0077] Technical solutions:
[0078] Such as Figure 3 ~ Figure 4 shown.
[0079] (1) Compound target protein binding data (positive sample), compound-target protein non-binding data (negative sample)
[0080] Compound target protein binding data (positive sample), compound-target protein non-binding data (negative sample). Among them, the positive sample is the target protein binding of the compound that humans have discovered in scientific research activities, usually selected from ChemSpider, PubChem, BindingDB, ZINC, ChEMBL...
Embodiment 2
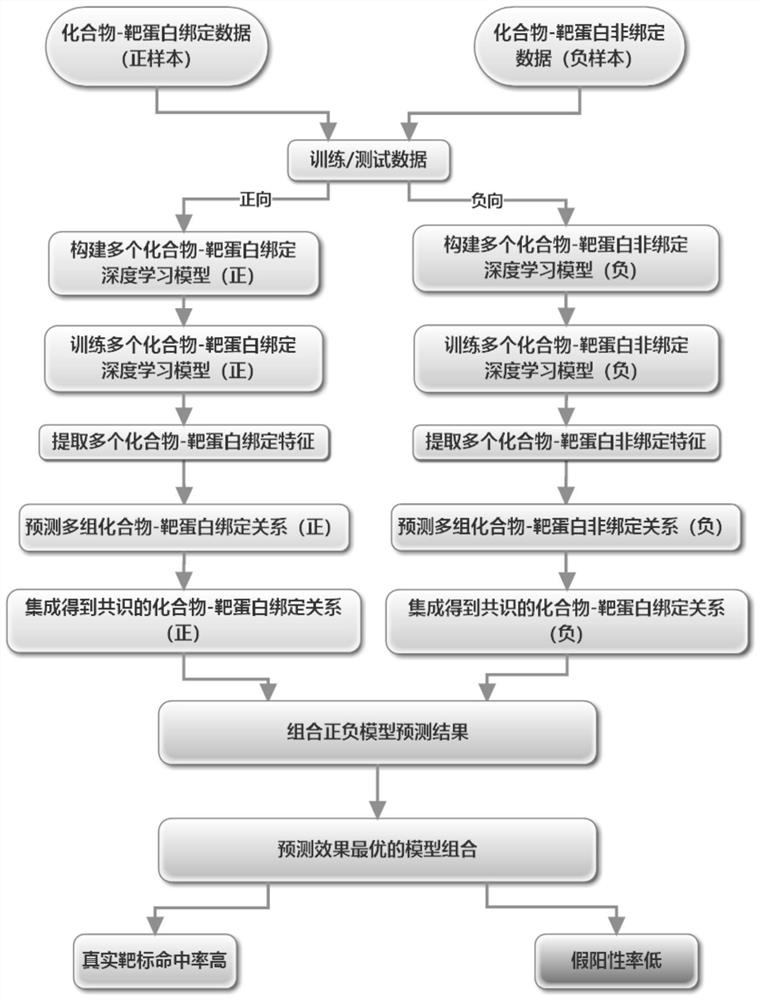
[0127] Multiple "recurrent neural networks (positive)" are connected in series with multiple "encoder-decoder neural networks (negative)" deep learning models to predict the binding relationship and system of compound target proteins.
[0128] Schematic diagram of the overall system structure: see Figure 5 .
[0129] System operating environment: same as "Embodiment 1".
[0130] (1) Obtain compound target protein binding / non-binding data (positive sample / negative sample)
[0131] The same as the corresponding part in "Example 1".
[0132] (2-1) Extract compound target protein binding / unbinding data (positive sample / negative sample) according to nine positive and negative sample ratios and synthesize the total data set, which is divided into training set, test set and verification set
[0133] With (2-1) part of " embodiment 1 " technical scheme.
[0134] (2-2) Build multiple different compound target protein binding / non-binding deep learning models (positive / negative mode...
Embodiment 3
[0160] Multiple "encoder-decoder neural networks (positive)" connected in series with multiple "recurrent neural network (negative)" deep learning models to predict the binding relationship and system of compound target proteins
[0161] Schematic diagram of the overall system structure: see Figure 6 .
[0162] System operating environment: same as "Embodiment 1".
[0163] Technical solutions:
[0164] (1) Obtain compound target protein binding / non-binding data (positive sample / negative sample)
[0165] The same as the corresponding part in "Example 1".
[0166] (2-1) Extract compound target protein binding / unbinding data (positive sample / negative sample) according to nine positive and negative sample ratios and synthesize the total data set, which is divided into training set, test set and verification set
[0167] With (2-1) part of " embodiment 1 " technical scheme.
[0168] (2-2) Build multiple different compound target protein binding / non-binding deep learning model...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com